Figure 5.
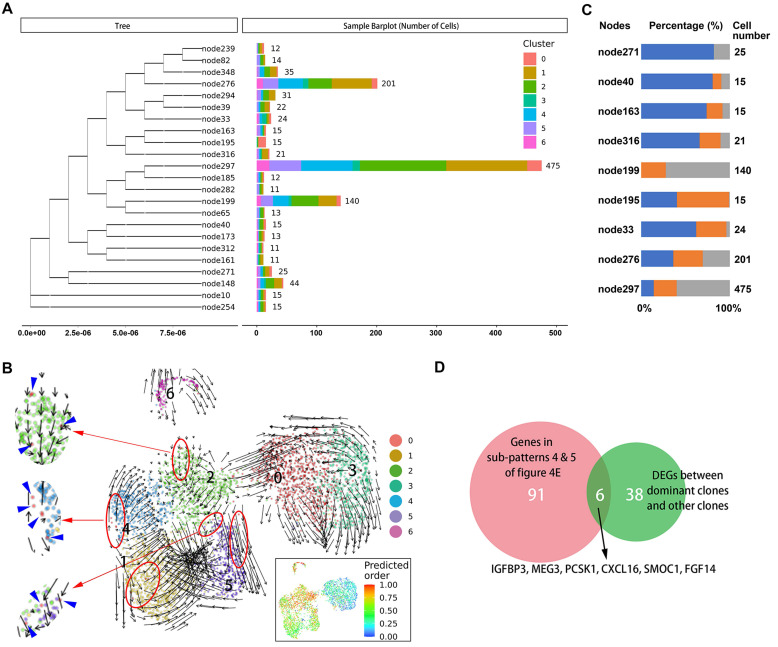
A. Branches of the mitochondrial phylogenetic tree of malignant cells. Cell numbers in each branch, as well as the clusters information are shown. B. RNA velocities are visualized on the UMAP projection with clusters information of malignant cells. The inset plot (in the black box): CytoTRACE results were visualized on the UMAP projection, showing the predicted developmental order of cells; color key from blue to red indicates pseudo-time from initiation to end. Blue arrows highlighted the distributed cells of cluster 0. C. Components of cell source in main branches were estimated and summarized in the bar chart. Total number of malignant cells in each site [primary tumor 1 (T1), primary tumor 2 (T2), liver metastasis (H5)] was used as reference for normalizing the cell proportion of corresponding sites in each branch. Blue bar, primary tumor 1 (T1); orange bar, primary tumor 2 (T2); gray bar, liver metastasis (H5). D. Venn plot showing the intersection of genes steady increase during tumor progression and genes significantly dysregulated in dominant clones.