Table 2.
Highest scoring docked conformations of ACE2–SARS-CoV2 S spike glycoprotein complexed with different ligands
| Ligands | Molecular structure | ΔGa (kcal/mol) | ΔGb (kcal/mol) | H bonding Ligand–protein |
Non-covalent interactions Ligand–protein |
|
|---|---|---|---|---|---|---|
| ACE inhibitor (Sigma) |

|
− 10.0 ± 0.033 | − 9.2 ± 0.033 |
C=O(1)–Lys74(A) C=O(5)–Trp69(A) C=O(9)–Trp349(A) Arg(6)NH2–Met62(A) Arg(6)NH–Ser43(A) |
Pro(1)–Ser70(A) Pro(2)–Leu100(A) Ile(3)–Gln102(A) Pro(5)–Leu73(A) Pro(7)–Trp349(A) Trp(8)–Phe40/Tyr385(A) Cyc(10)-Ala348(A) |
|
| Remdesivir |
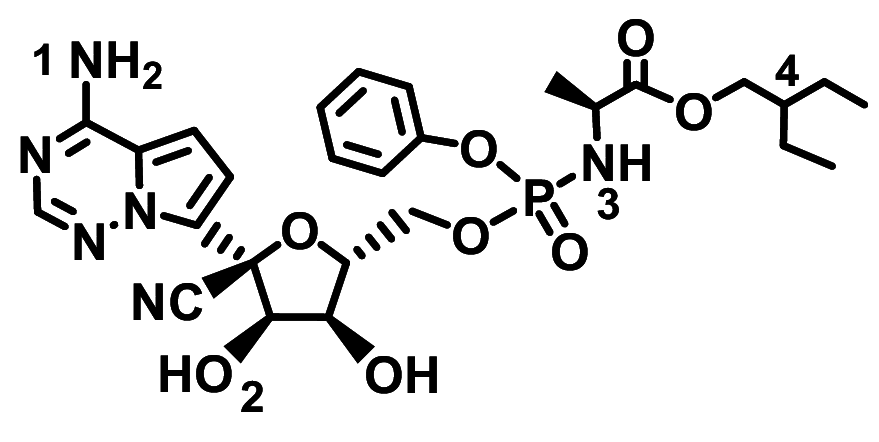
|
− 8.1 ± 0.15 | − 7.9 ± 0.070 |
NH2(1)–Glu398(A) NH(3)–Asp350(A) |
Alkyl(4)–Phe390/Phe40(A) Phe–Ser47(A) OH(2)–His378(A) |
|
| Hydoxychloroquine |

|
− 6.3 ± 0.09 | − 6.3 ± 0.22 | NH(1)–Tyr385(A) |
OH–Arg393(A) Pyr N–His378(A) N2–Arg393/Asp350(A) |
|
| Chloroquine |

|
− 6.1 ± 0.07 | − 6.1 ± 0.22 | NH(1)–Tyr385(A) |
OH–Arg393 Pyr N–His378(A) N2–Arg393/Asp350/Phe40(A) |
|
| Dexamethasone |
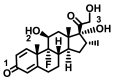
|
− 7.2 ± 0.00 | − 7.3 ± 0.00 | C=O(3)–Ser47/Trp349(A) |
OH(2)–Asp350(A) OH(3)–Asp382/Tyr385(A) |
|
| Losartan |
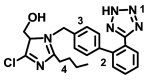
|
− 7.6 ± 0.19 | − 7.2 ± 0.088 | None |
OH–His378/Ala348(A) Het(1)–Asn394(A) Phe(2)–Phe40/Arg393(A) Alkyl(4)–Try349(A) |
|
| Olmesartan |
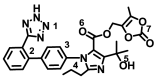
|
− 7.7 ± 0.033 | − 7.4 ± 0.033 |
N(1)–Asp350(A) C=O(6)–Ala348(A) |
Ph(2)–Trp349/Phe40(A) Alkyl(4)–Thr347(A) Cyc(7)–His401(A) |
|
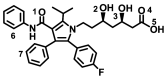
| Atorvastatin |
|
− 8.1 ± 0.067 | − 8.0 ± 0.058 |
OH(3)–His378(A) OH(5)–Pro346(A) C=O()–Asp350(A) |
Ph(1)–Tyr385(A) Ph(F)–Trp349/Phe40(A) |
|
| Simvastatin |

|
− 6.5 ± 0.00 | − 6.7 ± 0.00 | OH–Asp382(A) |
Cyc(1)–Phe40(A) Cyc(2)–Trp69/Phe390(A) Cyc(3)–Asp350(A) |
|
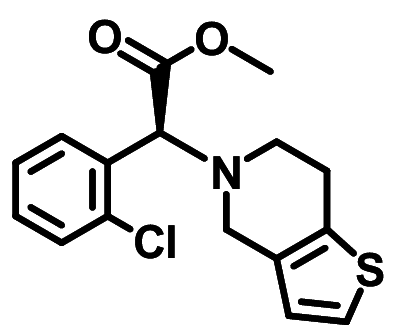
| Clopidogrel |
|
− 6.9 ± 0.00 | − 6.7 ± 0.067 | None |
Ph(Cl)–Phe40(A) Thiphenyl–Asp350(A) |
|
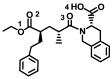
| Quinapril |
|
− 8.4 ± 0.033 | − 8.1 ± 0.23 | C=O(3)–Gln98(A) |
O(1)–Gln102(A)/Tyr202(A) OH(4)–Leu95(A) Bn–Asn194(A) Fused Ph–Leu95/Lys562(A) |
|
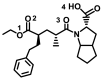
| Ramepril |
|
− 7.7 ± 0.14 | − 8.0 ± 0.14 | C=O(2)–Lys441(A) |
Bn–Phe438/Pro415(A) OH(4)–Ser405/Leu370(A) Cp–Leu410(A) O(1)–Asp292(A) |
|
| Benazepril |
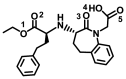
|
− 7.6 ± 0.10 | − 7.3 ± 0.27 | C=O(3)–Asp350(A) |
Fused Ph–Phe40/Trp349(A) C=O(2)–Arg393(A) Bn–Leu391(A) |
|
| Moexipril |

|
− 7.5 ± 0.00 | − 7.6 ± 0.088 |
C=O(2)–Try349 (A) C=O(4)–Try349(A) OH(4)–Asp350(A) |
O(5)–Ser47/Met62(A) Fused Ph–Phe40(A) Bn–Phe390(A) C=O(4)–Try349(A) EtO(1)–His378/His401(A) |
|
| Zofenopril |
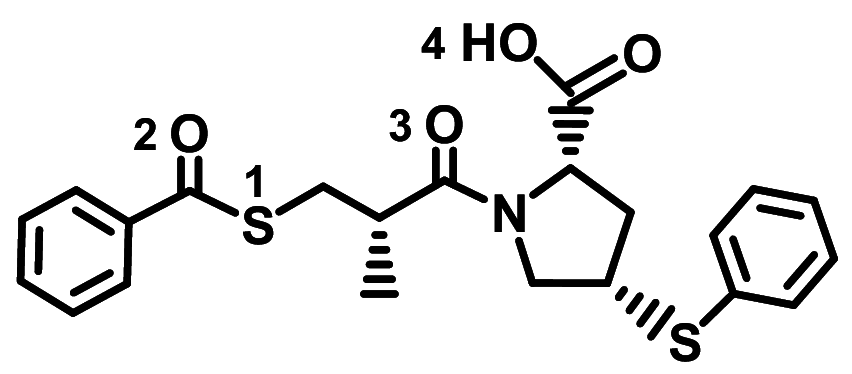
|
− 7.4 ± 0.033 | − 7.6 ± 0.12 | None |
PhS–Ser47/Ser44(A) C=O(3)–Asn394(A) OH(4)–Tyr385(A) PhC=O(2)–Arg393(A) |
|
| Enalapril |

|
− 7.1 ± 0.20 | − 7.4 | C=O(2)–Lys441(A) |
Bn–Phe438/Ile291/Met366(A) OH(4)–Ser405/Leu370(A) Pyr–Leu410(A) O(1)–Asp292/Asp367(A) |
|
| Fosenopril |

|
− 7.3 ± 0.00 | − 7.5 ± 0.11 | P=O–Asp350(A) |
iPr–His401(A) Ph–Tyr385/Phe40(A) Cyc–Thr347Trp349(A) P=O–Asp382(A) |
|
| MLN-4760 |
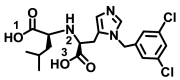
|
− 6.9 ± 0.12 | − 7.3 ± 0.067 |
OH(1)–Ala348(A) NH(2)–Ala348(A) OH(3)–Asp340(A) |
iBut–His401(A) Ph–Asp350/Arg393(A) |
|
| Perindopril |
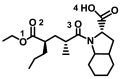
|
− 6.3 ± 0.00 | − 6.2 ± 0.15 | None |
OH(4)–Gln81/Asn103(A) C=O(3)–Gln98(A) Cyc–Leu95(A) O(1)–His195/Tyr196(A) |
|
| Nicotianamine (NAM) |

|
− 5.9 ± 0.088 | − 6.2 ± 0.14 |
C=O(5)–Trp566(A) OH(5)–Asn210(A) OH(1)–Gly205(A) NH(2)–Gly205(A) |
Cyc–Leu95(A) CycN–Lys562(A) NH(4)–Gln98(A) C=O(1)–Tyr196(A) |
|
| Captopril |
|
− 4.8 ± 0.00 | − 5.4 ± 0.033 | C=O(3)–Met270(A) |
SH–Thr276(A) Met–Phe274(A) OH(1)–Ala153(A) CycPro–Met270/Trp271(A) |
|
| N-(2-aminoethyl)-1-aziridine-ethanamine (NAAE) |

|
− 3.8 ± 0.12 | − 3.8 ± 0.033 | None |
Cyc–Glu435/Phe438(A) NH(2)–Ile291/Thr434(A) NH2(1)–Asn290/Asn437(A) |
|
ΔGa and ΔGb were determined using PDB ID:6M0J and PDB ID:6LZG, respectively. Ligand-protein interactions were derived from PDB ID:6M0J
A chain A or ACE2 receptor, E SARS-CoV-2 spike protein or chain E, Ph benzene ring, Pyr pyridine, Cyc cyclic chain, Bn benzyl ring, Cp cyclopentyl, Et ethyl, iPr isopropyl, iBut isobutyl
