Figure EV1. Excavation of the most conserved residues in APN globular domain, and the screening of candidate 5‐mer peptides based on DP and ORS cell proliferation.
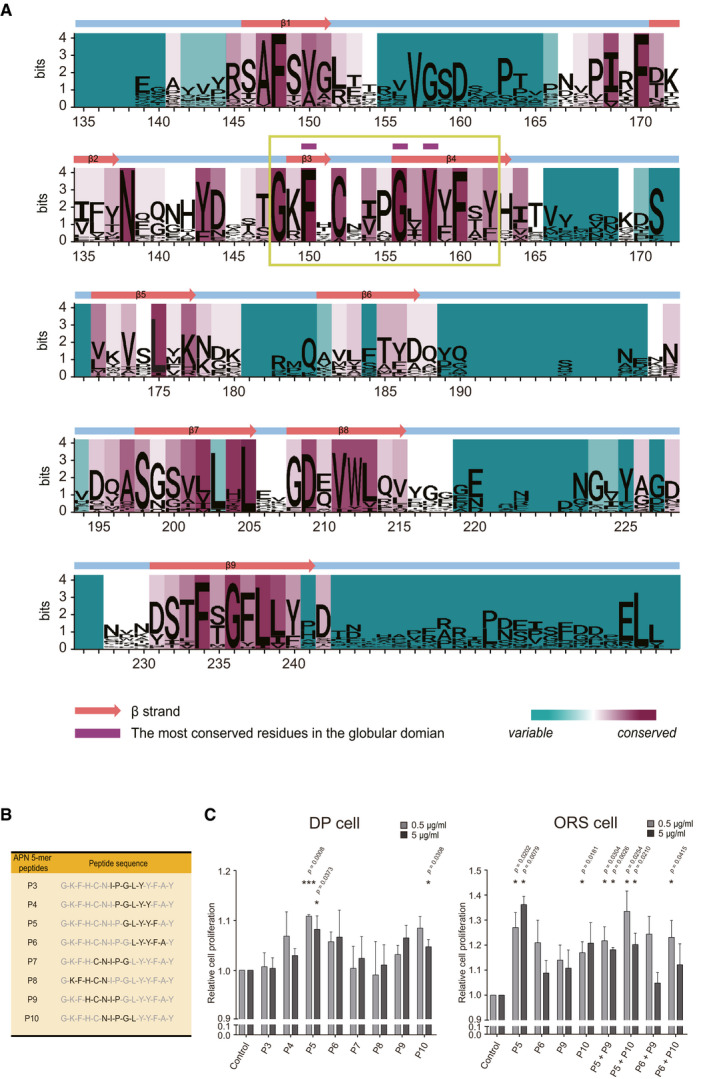
- Sequence logo landscape of MSA of APN globular domain. The residue numbers are based on the human APN amino acid sequence. In a given residue, the height of each letter (measured in bits) is proportionally scaled to the amount of information contributed based on Shannon entropy. The background color of each residue is determined by the degree of conservation of physicochemical properties (cyan to magenta). The most conserved residues (marked with purple tags) are located within a highly conserved sequence (designated in a green box).
- Candidate 5‐mer peptide sequences derived from a highly conserved sequence of APN globular domain. Each sequence is indicated in black.
- DP and ORS cells are used to screen the candidate 5‐mer peptides in terms of the cell proliferation effect (n = 3–5 biological replicates/group).
Data are presented as mean ± SEM. Statistical significance was determined using Paired t‐test (*P < 0.05 or ***P < 0.001 compared to the control group).
Source data are available online for this figure.
