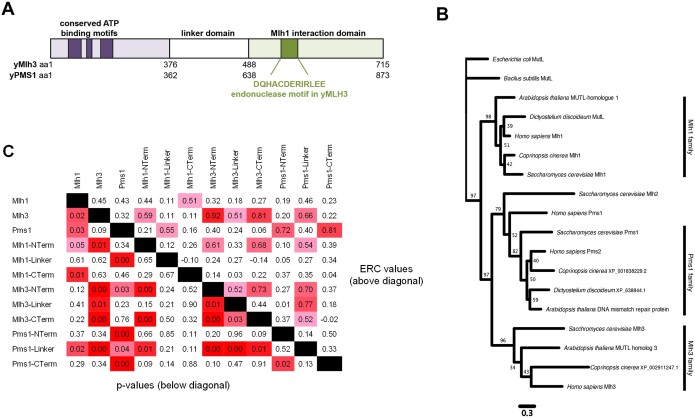
Figure 1.
Phylogenetic and ERC analysis of MLH proteins. (A) Cartoon depictions of the Mlh1-Mlh3 and Mlh1-Pms1 complexes, with the N-terminal ATP binding and C-terminal endonuclease/Mlh1 interaction domains separated by intrinsically disordered linker domains. The amino acid locations of the domains in yeast Mlh3 and Pms1 are shown. (B) Phylogenetic analysis of divergent eukaryotes indicates that MLH gene duplications occurred in early eukaryote history before the divergence of the lineages leading to plants, fungi, animals, and amoebozoa. The scale bar indicates the number of changes per amino acid site, and bootstrap support values out of 100 are found near each node. Escherichia coli MutS and B. subtilis MutL were used to root the tree. The branching pattern supports the earliest divergence of the Mlh1 family followed by splits to form the Mlh2, Mlh3, and Pms1 families. Uniprot, NCBI, and RefSeq identifications for the genes used to make the tree can be found in Supplementary Table S4. (C) ERC is elevated between domains of MLH proteins. This pairwise matrix shows all comparisons between the full-length Mlh1, Mlh3, and Pms1 proteins and their ATP binding (abbreviated as Nterm), linker, and endonuclease/MLH interaction (abbreviated as Cterm) domains defined in (A). The domain boundaries of the 18 fungal species were obtained from phylogenetic alignments with the S. cerevisiae proteins. ERC values are above the diagonal and empirical P-values are below. The colors of ERC cells range from pink at values of 0.5 to red at 1.0. P-value cells are pink at 0.05 and become red as they approach zero.