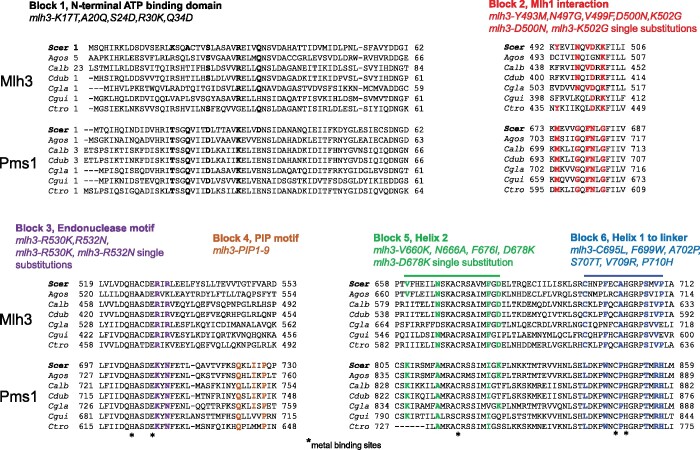
Figure 2.
Mutations made in MLH3 to revert back to conserved PMS1 sequences. Six blocks of mutations were made in Mlh3; Block 1-ATP binding, Block 2-Mlh1 interaction, and Blocks 3 to 6-Endonuclease/PCNA interaction. The multiple amino acid substitutions are shown for each block as well as single substitutions that were made in each region. For Blocks 1, 2, 5, and 6, Multi-Relief and Sequence-Harmony algorithms were used to identify functionally specific residues in Pms1 (Materials and Methods; adapted from Furman et al. 2021). Briefly, Mlh3 and Pms1 amino acid sequences from 34 different fungal species were aligned and presented in multi-Harmony. Shown are alignments of the regions showing seven fungal species (S. cerevisiae-Scer, Ashbya gossypii-Agos, Candida albicans-Calb, Candida dubliniensis-Cdub, Candida glabrata-Cgla, Candida guilliermondii-Cgui, Candida tropicalis-Ctro). The more complete lists of species alignments are shown in Supplementary Figure S4 legend. The N and C-terminal domains of Mlh3 were mapped onto the 3D structure of Mlh1-Pms1, and four amino acid clusters were identified for substitution analysis (Manning et al. 2008; Brandt et al. 2010). Block 3 spans the endonuclease motif found in Pms1 and Mlh3 (Supplementary Figure S3D). Block 4 contains the QXLXXP motif important for interactions with PCNA (PIP), which is highly conserved in the Pms1 sequences (>94% identity; Genschel et al. 2017) but is absent in Mlh3 sequences. Nine PIP mutations were made as shown in Supplementary Figure S3D and Tables 1 and 2. The asterisks indicate highly conserved metal-binding residues (H703, E707, C817, C848, and H850) in yeast Pms1, which form the endonuclease active site (Gueneau et al. 2013).