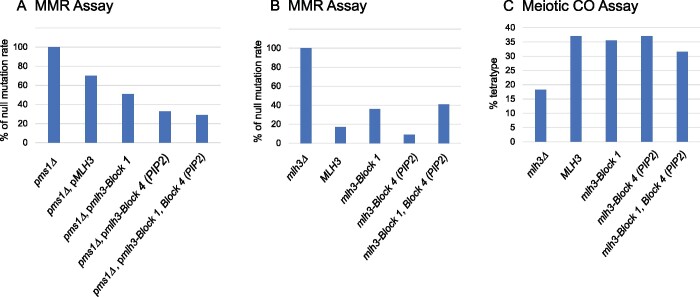
Figure 3.
Block 1 and Block 4 mlh3 mutations can alleviate the pms1Δ MMR defect. (A) Mutation rates of mlh3-Block 1 and mlh3-Block 4 single‐ and double‐mutant alleles expressed on 2µ plasmids in pms1Δ strains were determined in the lys2-A14 assay as described in Materials and Methods. Rates are shown as a percentage of the corresponding pms1Δ. (B) Mutation rates of strains containing mlh3 constructs integrated at the MLH3 locus in PMS1 strains. Rates are shown as a percentage of an mlh3Δ strain. (C) Meiotic crossover phenotypes, expressed as % tetratype, for diploid strains containing mlh3 constructs integrated at the MLH3 locus. For (A), MMR rates were normalized from data presented in Table 2, with the lines from the table where the data were obtained indicated with PMS1, ARS-CEN corresponding to line 1. Data were obtained from pms1Δ strains containing an empty 2µ vector-line 4 (set to 100%), pMLH3-line 48, pmlh3-Block 1-line 10, pmlh3-Block 4 (PIP2)-line 23, and pmlh3 Block 1, Block 4 (PIP2)-line 37. For (B), MMR rates were normalized from data presented in Table 1 (lines indicated, with MLH3 corresponding to line 1) and obtained as follows: mlh3Δ-line 2 (set to 100%), MLH3-line 1, mlh3-Block 1-line 13, mlh3-Block 4 (PIP2)-line 24, and mlh3-Block 1, Block 4 (PIP2)-line 32. For (C), CO data were obtained from data presented in Table 1 (lines indicated) as follows: mlh3Δ-line 2, MLH3-line 1, mlh3-Block 1-line 13, mlh3-Block 4 (PIP2)-line 24, and mlh3-Block 1, Block 4 (PIP2)-line 32.