Abstract
Nucleotide composition (GC content) varies across bacteria species, genome regions, and specific genes. In Xylella fastidiosa, a vector-borne fastidious plant pathogen infecting multiple crops, GC content ranges between ∼51-52%; however, these values were gathered using limited genomic data. We evaluated GC content variations across X. fastidiosa subspecies fastidiosa (N = 194), subsp. pauca (N = 107), and subsp. multiplex (N = 39). Genomes were classified based on plant host and geographic origin; individual genes within each genome were classified based on gene function, strand, length, ortholog group, core vs accessory, and recombinant vs non-recombinant. GC content was calculated for each gene within each evaluated genome. The effects of genome and gene-level variables were evaluated with a mixed effect ANOVA, and the marginal-GC content was calculated for each gene. Also, the correlation between gene-specific GC content vs natural selection (dN/dS) and recombination/mutation (r/m) was estimated. Our analyses show that intra-genomic changes in nucleotide composition in X. fastidiosa are small and influenced by multiple variables. Higher AT-richness is observed in genes involved in replication and translation, and genes in the leading strand. In addition, we observed a negative correlation between high-AT and dN/dS in subsp. pauca. The relationship between recombination and GC content varied between core and accessory genes. We hypothesize that distinct evolutionary forces and energetic constraints both drive and limit these small variations in nucleotide composition.
Keywords: Xylella fastidiosa, nucleotide composition, GC content, information storage and processing (ISP)
Introduction
The relevance of nucleotide composition (GC content) in genome evolution has been well established from a genomics (Arndt et al. 2005; Amit et al. 2012; Šmarda et al. 2014; Mugal et al. 2015; Almpanis et al. 2018), ecological (Bolhuis et al. 2006; Šmarda et al. 2014; Luo et al. 2015), and biological perspective (Mann and Chen 2010; Udaondo et al. 2016; Bohlin et al. 2017; Du et al. 2018; Castillo et al. 2019a). Specifically in proteobacteria, whole-genome GC content varies between 17% and 75% (Brocchieri 2014), and multiple evolutionary mechanisms have been proposed to explain this variation. One mechanism is GC-biased gene conversion (gBGC), which refers to a repair bias favoring GC over AT alleles during recombination (Galtier et al. 2001). This mechanism describes how highly recombinant genome regions, or organisms where recombination is more frequent, tend to be more GC-rich (Lassalle et al. 2015; Romiguier and Roux 2017). Specific nucleotide compositions can also be favored by natural selection. For example, the energetic cost of synthesizing A/T nucleotides is considered lower than that of synthesizing G/C nucleotides (Du et al. 2018). Hence, nucleotide composition differences in energetically constrained environments are explained by the energetic limitations to nucleotide synthesis (Li et al. 2015). Genetic GC content is linked to gene expression; in bacteria higher GC content is correlated with higher expression (Zhou et al. 2014; Chen et al. 2016) and fitness (Raghavan et al. 2012). Intra-genic changes in GC content have also been associated with mRNA stability; GC content is reduced near the start codon of protein-coding genes, particularly in those with higher average GC content, as a mechanism to facilitate protein translation (Gu et al. 2010). Finally, an important factor affecting GC-content is mutation bias. With some exceptions (McCutcheon et al. 2009; Dillon et al. 2015), mutation in bacterial genomes is AT-biased (Hershberg and Petrov 2010). It is believed that this is a result of a bias toward transitions at fourfold degenerate sites (Hildebrand et al. 2010) with C to T and G to A transitions being consistently favored over T to C and A to G transitions, even in GC-rich genomes (Hershberg and Petrov 2010). The bias is particularly evident in obligate symbionts due to loss of DNA repair genes (Moran et al. 2008) and small effective population size (Balbi et al. 2009). Moreover, balances between mutation trends and natural selection have a strong impact on nucleotide composition. For example, the GC content of substituted bases (sbGC) has been found to be GC biased compared with the GC content of the core genome in most microbial genomes (except for those highly GC-rich). This trend has been proposed to be linked to natural selection countering the C/G to T/A transitional bias universally observed in microbial genomes (Bohlin et al. 2018).
Overall, no single mechanism is likely to fully explain changes in nucleotide composition. Instead, GC content variations likely stem from interactions and balances among adaptive and non-adaptive forces (e.g. mutation vs selection). As a result, distinct genes, gene/genome regions, genomes, and populations can achieve unique GC content values (Botzman and Margalit 2011). Studying changes in GC content can highlight general trends, pinpoint taxa not following those trends, and illustrate evolutionary mechanisms relevant for a group. In this regard, while it is known that bacterial genome composition follows unique evolutionary trends; most analyses have been largely skewed toward species of clinical and veterinary interest (Lassalle et al. 2015; Bohlin et al. 2017). In terms of plant-associated bacteria, only 0.1% of the estimated 3 million taxa have been studied (Ingram 2002), with pathogens affecting the agricultural and forestry industries being overrepresented (Mansfield et al. 2012; Almeida 2018). These studies are commonly aimed to describe aspects of biogeography, host-pathogen adaptive potential, or pathogen management. Because of this, there is little to no research characterizing changes in plant-pathogen genome structure and nucleotide composition, despite its undisputed evolutionary role.
Both intra- and inter-genomic GC content variations have been identified in specific bacterial plant symbionts. Changes in intra-genomic GC content are indicative of a highly flexible genome in Xanthomonas campestris (Thieme et al. 2005). Also, similarities between genomic GC content within Erwinia and Enterobacter suggest that, contrary to expectation, nucleotide composition is preserved across the free living and obligate symbiont lifestyles (Estes et al. 2018). These are indicators that GC content in bacterial plant-associated bacteria has unique intra- and inter-genomic trends worthy of exploring. However, there are too few studies focused on the matter to highlight any pattern. Creating a comprehensive analysis in GC content variation is unfeasible, but this knowledge gap could be breached by characterizing pivotal groups. In this regard, Xylella fastidiosa, an emerging vector-borne plant pathogen with an expanding host and geographic range (EFSA 2018), represents a key study system due to its obligate colonization of plant and insect vector hosts.
Compared with its closest relative (i.e. Xanthomonas spp.), X. fastidiosa has undergone substantial genomic and biological changes. The X. fastidiosa genome is half the size of Xanthomonas spp. and characterized by the loss of specific metabolic functions and a slow growth rate (Gerlin et al. 2020). Unlike Xanthomonas spp., X. fastidiosa is transmitted by xylem sap-feeding insects (Vicente and Holub 2013; Cornara et al. 2017; Overall and Rebek 2017). Multiple studies have highlighted how changes in lifestyle influence genomic nucleotide composition (Foerstner et al. 2005; Merhej et al. 2009; Mann and Chen 2010; Dutta and Paul 2012; Aslam et al. 2019). In particular, lower GC content can result from drops in effective population size (Lassalle et al. 2015) and nutritional limitations (Mann and Chen 2010), both of these are variables found in vector-borne pathogens. In addition, X. fastidiosa has been introduced to multiple naïve crop populations (Schuenzel et al. 2005; Nunney et al. 2013; Landa et al. 2020) where it is hypothesized to evolve clonally (Ramazzotti et al. 2018; Sicard et al. 2018). This geographic component could affect nucleotide composition, since previous studies have highlighted how stronger AT-biases are observed in clonal bacterial populations due to the relaxation natural selection (Hershberg and Petrov 2010). For these reasons, characterizing GC content in X. fastidiosa might aid in better understanding of how evolutionary and biological forces shape the genomes of plant-associated bacteria.
The available X. fastidiosa genomes reported an average GC content of 51–52% (Simpson 2000; Castillo et al. 2019b); however, no study has evaluated inter-genic variations in GC content, defined the evolutionary forces shaping nucleotide composition, or compared these trends with those of other phytopathogens. We examined gene-specific GC content variations in subsp. pauca, subsp. multiplex, and subsp. fastidiosa. We evaluated if changes in GC content were associated with ecological (i.e. host plant, geographic source, etc.), functional (i.e. clusters of orthologous group [COGs]), evolutionary (i.e. selection, core/accessory, or recombinant/non-recombinant genes), or genetic variables (i.e. gene position, gene length, gene strand, etc.). Finally, we compared the GC content of X. fastidiosa with four other plant pathogens: Xanthomonas citri, X. campestris, Xanthomonas oryzae pv. oryzae, and Xanthomonas oryzae pv. oryzicola (which are closely related to X. fastidiosa), and the more distant Agrobacterium tumefaciens.
Materials and methods
Whole-genome sequences from worldwide X. fastidiosa isolates, and publicly available X. citri, X. camprestri, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, and A. tumefaciens isolates
The following study encompasses 340 X. fastidiosa whole-genome sequences. Isolates were collected from infected plant material in diverse geographic regions. Isolates belong to subsp. fastidiosa (N = 194), subsp. multiplex (N = 39), and subsp. pauca (N = 107). The number of isolates varied among geographic locations and infected plant hosts. Raw data have been made publicly available (Supplementary Table S1). Most isolates were sequenced using Illumina HiSeq2000; however, five subsp. fastidiosa isolates were sequences using both Illumina HiSeq2000 and PacBio (Castillo et al. 2020); and five subsp. pauca isolates were sequenced from total plant DNA (Sicard et al. in preparation). Samples were sequenced at the University of California, Berkeley Vincent J. Coates Genomics Sequencing Laboratory (California Institute for Quantitative Biosciences; QB3). The quality of the genome assemblies varied but coverage remained above ∼59×.
Details on genome assembly and annotation protocols have been provided in previous studies (Castillo et al. 2019b, 2020; Landa et al. 2020). Briefly, the quality of raw paired FASTQ reads was evaluated using FastQC (Andrews and Wingett 2018) and visualized using MultiQC (Ewels et al. 2016). Following, Seqtk v1.2 (https://github.com/lh3/seqtk) and cutadapt v1.14 (Marcel 2011) were used to remove low-quality reads and adapter sequences, respectively. Pre-processed reads were assembled de novo with SPAdes v3.13 (Bankevich et al. 2012; Nurk et al. 2013). Assembled contigs were reordered with Mauve’s contig mover function (Rissman et al. 2009) using complete publicly available assemblies as references (Temecula1 (GCA_000007245.1) for subsp. fastidiosa, 9a5c (ASM672v1) for subsp. pauca, and M12 (ASM1932v1) for subsp. multiplex). Finally, genomes were annotated using Prokka (Seemann 2014). Contamination was suspected on subsp. fastidiosa isolate XF70 from Costa Rica and removed by mapping FASTQ reads to its closest phylogenetic relative as described by Castillo et al. (2020). In the case of isolates obtained from total plant DNA, QC FASTQ reads were mapped to the Olea europaea genome assembly (GCA_900603015.1) with bowtie2 v2.3.4.1 (Langmead and Salzberg 2012). A SAM file of paired unmapped reads was created using the −f 12 and −F 256 flags in Samtools v1.8 (Li et al. 2009) and subsequently converted to sorted BAM. Bedtools v2.26.0 (Quinlan and Hall 2010) was used to convert the sorted BAM file into FASTQ files. The host-removed reads were then assembled with SPAdes v3.13 as described previously. In addition, publicly available whole-genome assemblies for A. tumefaciens (N = 12), X. oryzae pv. oryzae (N = 75), X. oryzae pv. oryzicola (N = 13), X. campestris (N = 14), and X. citri (N = 68) were obtained from NCBI and re-annotated with Prokka. Metadata for these isolates have also been included in Supplementary Table S1.
Pan-genome analysis of phytopathogen groups
Roary v3.11.2 (Page et al. 2015) was used to calculate the size of the core (genes shared between 99 and 100% strains), soft-core (genes shared between 95 and 99% strains), shell (genes shared between 15 and 95% strains), and cloud (genes shared between <15% strains) genomes for each X. fastidiosa subspecies, A. tumefaciens, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. campestris, and X. citri. The soft-core, shell, and cloud genomes were compiled into the accessory genome (genes shared between <99% strains). Individual genome assemblies and annotation files were used to mine gene sequences within each genome. Individual genes were also classified regarding their number of orthologs. Finally, all individual genes were categorized based on their COGs (Tatusov 2000). Each gene was classified by its specific COG function (e.g. translation and ribosomal structure and biogenesis, transcription, nuclear structure, cell motility, lipid transport, and metabolism, etc.) and by the main categories to which these functions belong [i.e. Metabolism (M), Information Storage and Processing (ISP), and Cellular Processes and Signaling (CPS)]. Individual genes without defined COG (e.g. hypothetical proteins) were assigned to the Poorly characterized (P) category. Individual genes belonging to two or more functional classes were grouped as Multiple categories (MU). Approximately 30–40% of individual genes were classified within the P category.
Recombination detection and estimation of global genetic diversity in core alignments
Roary was used to create core genome alignments for each X. fastidiosa subspecies, A. tumefaciens, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. campestris, and X. citri. The global measure of genetic diversity (π) was calculated for the core genome alignment of each phytopathogen using the R package “PopGenome” (Pfeifer et al. 2014). Nucleotide diversity (π) measures the average number of nucleotide differences per site in pairwise comparisons among DNA sequences. This measurement has been used to characterize X. fastidiosa populations (Vanhove et al. 2019, 2020; Castillo et al. 2020), and thus, can be easily contrasted to earlier studies.
FastGEAR (Mostowy et al. 2017) was used with default parameters to identify lineage-specific (ancestral) and strain-specific (recent) recombinant segments in X. fastidiosa subspecies core genome alignments. A custom python script was used to find individual core genes contained entirely within recombinant segments. Core genes found entirely within recombinant regions were henceforth classified as “recombinant genes,” and the remaining genes were classified as “non-recombinant genes.” The correlation between GC content and the recombination/mutation rate (r/m) was also evaluated for individual gene alignments. Briefly, Roary was used to identify individual ortholog groups within each X. fastidiosa subspecies. The detected ortholog groups were programmatically aligned using MACSE v2 (Ranwez et al. 2018). The corresponding Maximum Likelihood (ML) trees were then built with RAxML (Stamatakis 2014). All trees were built using the GTRCAT substitution model and tree topology and branch support were assessed with 100 bootstrap replicates. Each gene alignment and tree were then used as input for ClonalFrameML (Didelot and Wilson 2015). A subspecies-wide Pearson correlation coefficient was performed between GC content and the log10 transformation of r/m.
Estimation of gene-specific GC content and statistical analysis
A custom python script was used to calculate gene-specific GC content values for each gene within individual X. fastidiosa, A. tumefaciens, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. campestris, and X. citri genomes. GC content in the third or wobble position (GC3) was also calculated for each gene within individual X. fastidiosa subspecies. The length of each gene was calculated based on its start and end positions. Variables were assigned as gene functional class, gene size, gene strand, accessory/core gene, number of orthologs, and recombinant/non-recombinant gene (gene-dependent); and geographic location and plant host (genome-dependent). Certain variables known to affect genomic GC content were not studied here. For example, large scope RNAseq data were not available at the time of analysis, precluding any assessment of gene expression levels and their relationship to GC content. It is expected that this type of data will become available in the future. Similarly, most of the whole genome sequences used have been assembled to the contig level, and thus, it was not possible to estimate gene position in a full chromosome. In this regard, gene position was obtained for three finished representative whole-genome assemblies: Temecula1 (GCA_000007245.1) for subsp. fastidiosa, 9a5c (ASM672v1) for subsp. pauca, and M12 (ASM1932v1) for subsp. multiplex. A synteny analysis among the assemblies was conducted using CoGe (https://genomevolution.org/coge/ [last accessed on March 19th, 2021]).
A mixed-effect ANOVA was used to evaluate the statistical contributions of gene-dependent and genome-dependent variables in the nucleotide composition of individual genes. The Genome ID and genome GC content were used as a random slope and the gene ortholog group identified by Roary was used as a random effect. The model was used to estimate marginal means for gene-specific GC content (marginal-GC). In other words, the marginal-GC content representing the estimated GC content values averaged across all variable levels of the linear regression model.
Correlation between genic GC content with dN/dS values
Orthologous genes were identified, aligned, and their ML trees were constructed as described in the “Recombination detection and estimation of global genetic diversity in core alignments” section of the Materials and methods. The relationship between GC content and gene-wide signs of selection was estimated. The rate of non-synonymous over synonymous substitutions (dN/dS) was programmatically calculated for each gene alignment using BUSTED (Kosakovsky Pond et al. 2005; Murrell et al. 2015). Briefly, BUSTED determines if there is at least one positively selected site across the alignment and in any branch of the phylogeny. The subspecies-wide Pearson’s correlation coefficient was calculated between GC content and the Tukey’s Ladder of Powers transformation of dN/dS values.
Data availability
Raw sequence files are available upon request. Published sequence data are available at GenBank; the accession numbers are listed in Supplementary Table S1. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14067449.
Results
GC content variations are more readily observed within the accessory genome
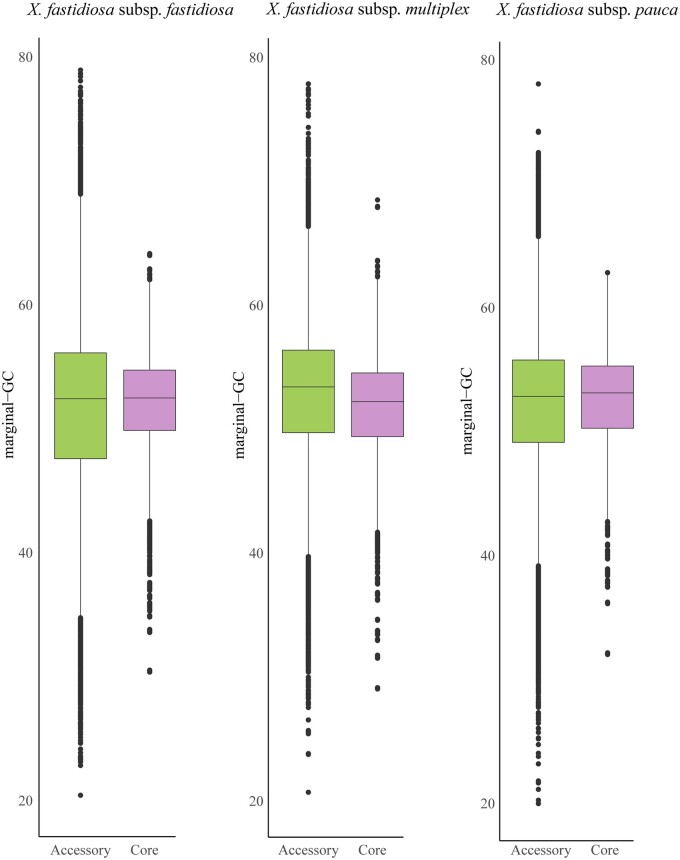
Core and accessory genome sizes varied within X. fastidiosa subspecies and in the other phytopathogens examined (Table 1). The standard deviation of genic GC content was lower in core genes compared with accessory genes (Figure 1 and Table 2). Gene-specific GC content also varied within accessory genome components for all three subspecies (Table 2). Specifically, genes in the shell genome had higher GC content variation compared with soft-core and cloud genomes in subsp. fastidiosa and subsp. pauca (Supplementary Figure S1). It should be noted that the shell genome has the widest range in relation to the percentage of strains sharing a given gene (15–95%). Therefore, it can potentially cover a wider range of gene presence/absence variations compared with the soft-core (genes shared by 95–99% of strains) and the cloud genomes (genes shared by <15% of strains).
Table 1.
Number of genes within each pan-genome component
| Phytopathogen | N | Genome component |
|||
|---|---|---|---|---|---|
| Core | Soft-core | Shell | Cloud | ||
| Xylella fastidiosa subsp. fastidiosa | 194 | 1,488 | 297 | 849 | 7,644 |
| Xylella fastidiosa subsp. multiplex | 39 | 1,006 | 226 | 2,152 | 5,982 |
| Xylella fastidiosa subsp. pauca | 107 | 1,244 | 470 | 1,100 | 6,513 |
| Xylella oryzae pv. oryzae | 75 | 2,460 | 311 | 3,812 | 4,824 |
| Xylella oryzae pv. oryzicola | 13 | 3,391 | 0 | 2,277 | 1,316 |
| Xylella citri | 68 | 1,084 | 1,887 | 2,444 | 11,027 |
| Xylella campestris | 14 | 3,372 | 0 | 1,664 | 1,987 |
| Agrobacterium tumefaciens | 12 | 1,638 | 0 | 9,774 | 11,009 |
N = number of genomes.
Figure 1.
Boxplot showing differences in marginal-GC content in core vs accessory genes in three X. fastidiosa subspecies.
Table 2.
(a) Mean GC content and standard deviation (sd) for genes in the core, accessory, and accessory genome components (soft-core, shell, and cloud genome) across subspecies. (b) F-test shows statistical significance of variance between groups
| Subspecies | Core | Accessory | Accessory components |
||
|---|---|---|---|---|---|
| Soft-core | Shell | Cloud | |||
| Xylella fastidiosa subsp. fastidiosa | 51.90 (4.37) | 51.56 (7.28) | 51.87 (6.01) | 51.31 (8.19) | 52.83 (9.16) |
| Xylella fastidiosa subsp. multiplex | 51.52 (4.78) | 52.49 (6.33) | 52.23 (5.36) | 52.47 (6.38) | 52.93 (7.39) |
| Xylella fastidiosa subsp. pauca | 52.51 (4.56) | 52.19 (6.17) | 52.23 (4.81) | 52.05 (7.73) | 52.32 (8.24) |
| Subspecies | Core vs accessory |
Soft-core vs shell | Shell vs cloud | Cloud vs Soft-core | |
| Xylella fastidiosa subsp. fastidiosa | F = 0.360 | F = 0.539 | F = 1.251 | F = 2.320 | |
|
P < 2.2 × 10−16*
|
P < 2.2 × 10−16* | P < 2.2 × 10−16* | P < 2.2 × 10−16* | ||
| Xylella fastidiosa subsp. multiplex |
F = 0.571 |
F = 0.706 | F = 1.343 | F = 1.903 | |
|
P < 2.2 × 10−16*
|
P < 2.2 × 10−16* | P < 2.2 × 10−16* | P < 2.2 × 10−16* | ||
| Xylella fastidiosa subsp. pauca |
F = 0.546 |
F = 0.386 | F = 1.135 | F = 2.935 | |
|
P < 2.2 × 10−16*
|
P < 2.2 × 10−16* | P < 2.2 × 10−16* | P < 2.2 × 10−16* | ||
Bold values correspond to p-values < 0.05. These values have also been marked with asterisk (*). *Statistically significant differences.
The nucleotide composition in cloud, shell, and soft-core genes was similar; however, there was a small and statistically significant decrease in GC content in soft-core genes relative to cloud genes (t = 14.866, P < 2.2 × 10−16 for subsp. fastidiosa; and t = 7.1555, P = 8.774 × 10−13 for subsp. multiplex) and in shell genes relative to cloud genes (t = 22.975, P < 2.2 × 10−16 for subsp. fastidiosa; t = 4.9127, P = 9.111 × 10−07 for subsp. multiplex; and t = 4.2695, P = 1.963 × 10−05 for subsp. pauca). However, this was not seen between the soft-core genes relative to cloud genes in subsp. pauca (t = 1.6353, P = 0.102). Trends in GC content variation based on explanatory variables are discussed below.
Apart from the variable “Host plant” and “Population,” most gene- and genome-dependent variables affected GC content (Table 3). This pattern was observed when using accessory and core gene classifications made by different pan-genome analysis programs [Roary vs GET_HOMOLOGUES (Contreras-Moreira and Vinuesa 2013)], when Hypothetical/Poorly characterized genes were removed from the dataset, and when Roary’s core genome threshold was reduced to 95% (Supplementary Table S2). GC3 variation followed the same trends (Supplementary Table S3). Statistically significant gene- and genome-dependent variables were subsequently plotted to evaluate if biological or ecological trends were present. In addition, the size effect (Cohen’s d) was calculated for the three statistically significant categorical variables: Accessory vs Core, DNA strand, and gene functional class (Table 4).
Table 3.
Mixed effect ANOVA results on gene-specific GC content
| Subspecies | GC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Accessory | Core | ||||||||
| Xylella fastidiosa subsp. fastidiosa | Variable | Chisq | Df | Pr(>Chisq) | Chisq | Df | Pr(>Chisq) | Chisq | Df | Pr(>Chisq) |
| Accessory/Core | 171.3993 | 1 | <2 × 10−16* | – | – | – | – | – | – | |
| Host | 162.6594 | 8 | <2 × 10−16* | 75.9588 | 8 | 3.17 × 10−13* | 80.8329 | 8 | 3.32 × 10−14* | |
| Gene length | 11749.963 | 1 | <2 × 10−16* | 7514.8866 | 1 | <2 × 10−16* | 417.0152 | 1 | <2 × 10−16* | |
| Strand | 914.5679 | 1 | <2 × 10−16* | 420.8673 | 1 | <2 × 10−16* | 934.1671 | 1 | <2 × 10−16* | |
| Populationa | 7.4435 | 7 | 0.3842 | 5.1146 | 7 | 0.646 | 0.7336 | 7 | 0.9981 | |
| Function | 1,801.8669 | 4 | <2 × 10−16* | 631.8232 | 4 | <2 × 10−16* | 10,113.0656 | 4 | <2 × 10−16* | |
| Number of orthologsb | 6,871.6529 | 189 | <2 × 10−16* | 2,462.0721 | 189 | <2 × 10−16* | – | – | – | |
| Xylella fastidiosa subsp. multiplex | Accessory/Core | 14.5632 | 1 | 0.0001 * | – | – | – | – | – | – |
| Host | 14.0509 | 15 | 0.5217 | 12.4927 | 15 | 0.6414 | 14.0804 | 15 | 0.5194 | |
| Gene length | 434.2259 | 1 | <2 × 10−16* | 255.1272 | 1 | <2 × 10−16* | 97.9301 | 1 | <2 × 10−16* | |
| Strand | 60.102 | 1 | 9.01 × 10−15* | 20.179 | 1 | 7.05 × 10−06* | 134.2912 | 1 | <2 × 10−16* | |
| Populationa | 5.5217 | 5 | 0.3556 | 6.0869 | 5 | 0.2979 | 1.3441 | 5 | 0.9303 | |
| Function | 265.9274 | 4 | <2 × 10−16* | 196.8562 | 4 | <2 × 10−16* | 81.2937 | 4 | <2 × 10−16* | |
| Number of orthologsb | 597.4762 | 37 | <2 × 10−16* | 354.634 | 37 | <2 × 10−16* | – | – | – | |
| Xylella fastidiosa subsp. pauca | Accessory/Core | 60.515 | 1 | 7.30 × 10−15* | – | – | – | – | – | – |
| Host | 5.153 | 5 | 0.3975 | 0.7541 | 5 | 0.9799 | 1.0277 | 5 | 0.9603 | |
| Gene length | 3,677.413 | 1 | <2 × 10−16* | 3,060.2776 | 1 | <2 × 10−16* | 758.9033 | 1 | <2 × 10−16* | |
| Strand | 567.02 | 1 | <2 × 10−16* | 457.0479 | 1 | <2 × 10−16* | 76.3242 | 1 | <2 × 10−16* | |
| Populationa | 2.298 | 3 | 0.5129 | 1.2317 | 3 | 0.7454 | 3.3509 | 3 | 0.3406 | |
| Function | 253.732 | 4 | <2 × 10−16* | 326.8554 | 4 | <2 × 10−16* | 118.7646 | 4 | <2 × 10−16* | |
| Number of orthologsb | 3,105.453 | 103 | <2 × 10−16* | 1,954.636 | 103 | <2 × 10−16* | – | – | – | |
Bold values correspond to p-values < 0.05. These values have also been marked with asterisk (*). *Statistically significant differences.
Refers to the geographic populations included in this study: California, Southeastern USA, Taiwan, Spain, Brazil, Italy, Costa Rica, and France.
Refers to the number of orthologues for each gene.
Table 4.
Cohen’s d of the accessory/core, DNA strand, and COG function classes
| Subspecies | Variables | Cohen’s d | 95% CI | |
|---|---|---|---|---|
| Xylella fastidiosa subsp. fastidiosa | Accessory/Core | −0.0571 | CI: (−0.063, −0.0513) | |
| Strand | 0.0392 | CI: (0.0333, 0.045) | ||
| Function | M vs CPS | −0.3202 | CI: (−0.3328, −0.3078) | |
| M vs ISP | −0.5112 | CI: (−0.5236, −0.4988) | ||
| ISP vs CPS | 0.2308 | CI: (0.2168, 0.2450) | ||
| Xylella fastidiosa subsp. multiplex | Accessory/Core | 0.167 | CI: (0.1526, 0.1814) | |
| Strand | 0.0504 | CI: (0.0364, 0.0645) | ||
| Function | M vs CPS | −0.345 | CI: (−0.3757, −0.3144) | |
| M vs ISP | −0.4868 | CI: (−0.5169, −0.4567) | ||
| ISP vs CPS | 0.1822 | CI: (0.1477, 0.2167) | ||
| Xylella fastidiosa subsp. pauca | Accessory/Core | −0.0553 | CI: (−0.0649, −0.0456) | |
| Strand | 0.0238 | CI: (0.0160, 0.0317) | ||
| Function | M vs CPS | −0.371 | CI: (−0.3879, −0.3542) | |
| M vs ISP | −0.5116 | CI: (−0.5284, −0.4949) | ||
| ISP vs CPS | 0.1874 | CI: (0.1684, 0.2064) | ||
Nucleotide composition varies between the leading and lagging DNA strand
The average number of individual genes in the lagging and leading strands was 1055 vs 1030 for subsp. fastidiosa, 1096 vs 1095 for subsp. pauca, and 1127 vs 1079 for subsp. multiplex. Overall, genes in the leading strand had lower marginal-GC content than those in the lagging strand in the core genome of subsp. multiplex and pauca, and the accessory genome of subsp. fastidiosa (Supplementary Figure S2). In addition to marginal-GC, GT was calculated for individual genes in the leading and lagging strand in all X. fastidiosa subspecies (Supplementary Figure S3). The goal was to establish if G and T nucleotides were enriched in either DNA strand (Lobry and Sueoka 2002). GT content was higher in the lagging strand of subsp. fastidiosa core genome and the core and accessory genomes of subsp. pauca.
ISP genes show lower GC content distribution
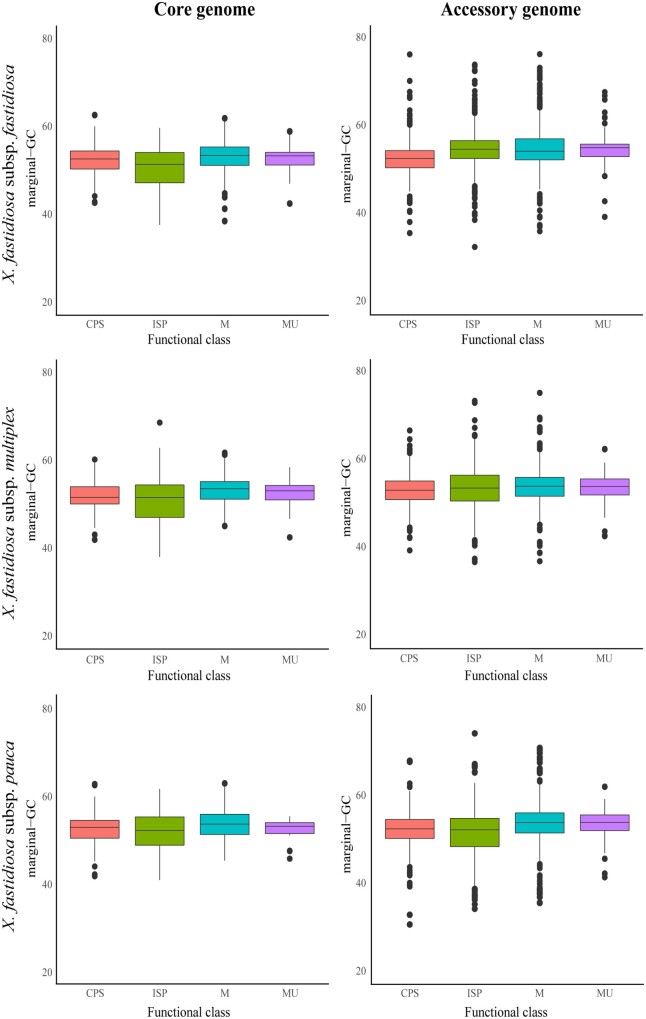
Gene functional class also affected genic GC content. The number of core and accessory genes from different functional groups varied within subspecies. The Metabolism (M) functional class was the most numerous, while the CPS and ISP functions had a similar number of genes (Table 5). There was no clear relation between GC content and gene number per function. In general, genes from the ISP class had lower marginal-GC content than genes from other functional groups (Figure 2) in the core genome of subsp. fastidiosa and subsp. multiplex, and in the accessory genome of subsp. pauca. Genes coding for ribosomal protein were the highest contributors to the lower marginal-GC content in the ISP functional class. Nonetheless, the variable “gene function” still had a significant effect even when these genes were removed from the dataset (Supplementary Table S4). After removal of ribosomal protein-coding genes, marginal-GC content in the CPS and ISP classes was similar in subsp. fastidiosa and subsp. multiplex core genes (Supplementary Figure S4). Notably, the marginal GC-content of M class genes was lower in the accessory genome, particularly in the case of subsp. fastidiosa. Genes from the Poorly characterized (P) category were removed from these visualizations.
Table 5.
χ 2 analysis showing statistically significant differences between the number of core and accessory genes from different COG functions within each Xylella fastidiosa subspecies
| Subspecies | Functional group | Accessory sum | Core sum |
|---|---|---|---|
| Xylella fastidiosa subsp. fastidiosa | Cellular Processes and Signaling (CPS) | 9,214 | 28,475 |
| Information Storage and Processing (ISP) | 8,159 | 31,538 | |
| Metabolism (M) | 17,018 | 56,590 | |
| Multiple categories (MU) | 1,719 | 5,301 | |
| Poorly characterized (P) | 166,938 | 124,154 | |
| χ2 = 49,336, df = 4, P < 2.2 × 10−16* | |||
| Xylella fastidiosa subsp. multiplex | Cellular Processes and Signaling (CPS) | 3,086 | 3,132 |
| Information Storage and Processing (ISP) | 2,906 | 3,817 | |
| Metabolism (M) | 6,186 | 6,185 | |
| Multiple categories (MU) | 572 | 633 | |
| Poorly characterized (P) | 33,890 | 17,239 | |
| χ2 = 2,503.1, df = 4, P < 2.2 × 10−16* | |||
| Xylella fastidiosa subsp. pauca | Cellular Processes and Signaling (CPS) | 14,599 | 6,416 |
| Information Storage and Processing (ISP) | 14,937 | 6,823 | |
| Metabolism (M) | 27,773 | 12,422 | |
| Multiple categories (MU) | 2,728 | 1,043 | |
| Poorly characterized (P) | 136,883 | 25,835 | |
| χ2 = 7,591.9, df = 4, P < 2.2 × 10−16* | |||
Accessory sum, sum of accessory genes from all isolates within each subspecies; Core sum, sum of core genes from all isolates within each subspecies. Bold values correspond to p-values < 0.05. These values have also been marked with asterisk (*).
Statistically significant differences.
Figure 2.
Boxplot showing marginal-GC content distribution on different functional classes for three X. fastidiosa subspecies. Plots have been divided into core/accessory genes within each X. fastidiosa subspecies. Functional classes include ISP (green), Cellular Processes and Signaling (CPS, blue), Metabolism (M, red), and Multiple categories (MU, purple).
Gene function had a significant effect on genic GC content even when genes were classified based on their specific function (i.e. “Translation,” “Amino acid transport and metabolism,” “Defense mechanism,” etc.) (Supplementary Table S5). The relationship between GC content in core vs accessory genes varied across functions, but overall, accessory genes had higher marginal-GC content than core genes of the same function in both subsp. fastidiosa and subsp. multiplex. This was particularly evident within the ISP functional class, where functions associated with “Translation” and “Replication” had lower marginal-GC content. Differences in nucleotide composition across specific functions were less clear in subsp. pauca (Supplementary Figure S5).
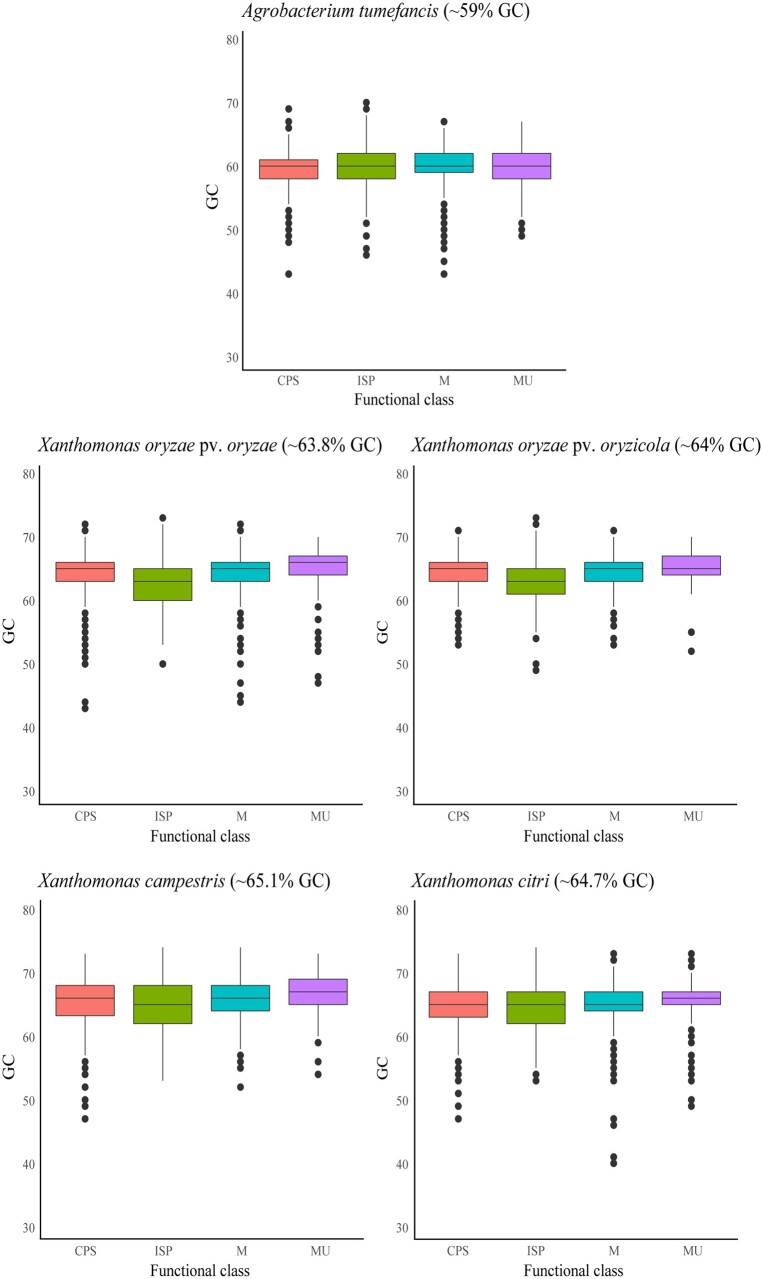
The distribution of marginal-GC content among functional classes was also evaluated in other plant pathogens (Supplementary Table S6): A. tumefaciens (283,876 SNPs and π = 0.059), X. oryzae pv. oryzae (45,414 SNPs and π = 0.007), X. oryzae pv. oryzicola (22,029 SNPs and π = 0.002), X. citri (111,198 SNPs and π = 0.012), and X. campestris (113,771 SNPs and π = 0.011). Neither X. citri, X. campestris, nor A. tumefaciens showed a clear separation in marginal-GC content between functional classes. On the other hand, in both X. oryzae pv. oryzae and X. oryzae pv. oryzicola genes from the ISP class had lower marginal-GC content compared with other functional classes, even after removal of ribosomal protein-coding genes (Figure 3). The trend was most prominent in X. oryzae pv. oryzae. Like in X. fastidiosa, functions associated with “Translation” and “Replication” maintained the lowest marginal-GC content.
Figure 3.
Boxplot showing marginal-GC content distribution on different functional classes in five plant-associated pathogens. Marginal-GC content distribution and average GC content is shown for each phytopathogen: A. tumefaciens, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. citri, and X. campestris. Functional classes include ISP (green), Cellular Processes and Signaling (CPS, blue), Metabolism (M, red), and Multiple categories (MU, purple).
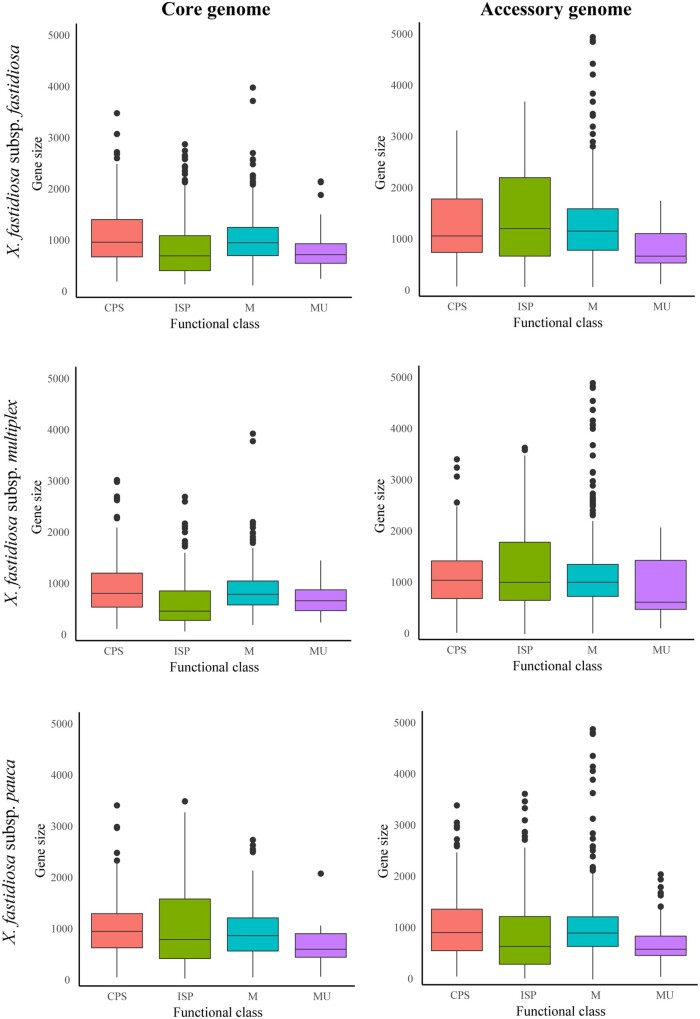
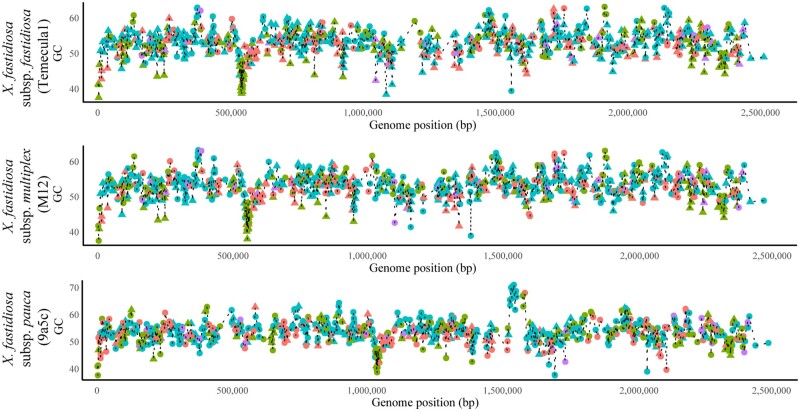
Genes from the ISP function were smaller than those from other functional groups (Figure 4); however, this difference was largely due to genes coding for ribosomal proteins (Supplementary Figure S6). Similarly, apart from a cluster of ribosomal protein-coding genes, there were no general trends between genic GC content and gene position (Figure 5). A synteny analysis of three complete genome assemblies representing each X. fastidiosa subspecies showed three chromosomal inversion events between the 9a5c strain (subsp. pauca), and the M12 (subsp. multiplex) and Temecula1 (subsp. fastidiosa) strains (Supplementary Figure S7).
Figure 4.
Boxplot showing gene size differences across gene functional classes for three X. fastidiosa subspecies. Plots have been divided into core/accessory genes within each X. fastidiosa subspecies. Functional classes include ISP (green), Cellular Processes and Signaling (CPS, blue), Metabolism (M, red), and Multiple categories (MU, purple).
Figure 5.
Line plot showing gene-specific GC content vs genes position across the length of three finished X. fastidiosa assemblies. The assemblies used are subsp. fastidiosa (strain Temecula 1), subsp. multiplex (strain M12), subsp. pauca (strain 9a5c). Core genes are shown with circles while accessory genes are shown with triangles. Functional classes include ISP (green), Cellular Processes and Signaling (CPS, blue), Metabolism (M, red), and Multiple categories (MU, purple).
Purifying selection is prevalent in genes from the core and accessory genome
Genic GC content and the Tukey’s Ladder of Powers transformation of dN/dS were significantly correlated for individual gene alignments of subsp. pauca [r(1930) = −0.0558, P = 0.0142], but not for subsp. multiplex [r(3869) = −0.0281, P = 0.081] and subsp. fastidiosa [r(3723) = −0.0247, P = 0.1316]. The significant correlation was small. Most individual gene alignments showed signs of gene-wide purifying selection, with only a small proportion having a dN/dS > 1 (subsp. multiplex 547/3872, subsp. fastidiosa 442/3726, and subsp. pauca 226/1933). Most of the genes under positive selection in each subspecies (subsp. multiplex 450/556, subsp. fastidiosa 393/442, and subsp. pauca 162/227) were classified in the P group. In subsp. fastidiosa, 12 genes belonged to the CPS class, 15 to the ISP class, and 19 to the M class; in subsp. multiplex, 23 genes belonged to the CPS class, 27 to the ISP class, and 50 to the M class; and in subsp. pauca, 12 genes belonged to the CPS class, 12 to the ISP class, and 38 to the M class (Supplementary Table S7).
There are no clear trends in GC content between recombinant and non-recombinant genes
For all analyzed X. fastidiosa subspecies, intra-subspecific recombination was pervasive across the length of the core genome alignment and equally frequent among functional groups (Table 6). As a general trend, marginal-GC content was lower in recombinant than non-recombinant genes (Supplementary Figure S8). Core genes within non-recombinant regions had similar lengths, while recombinant genes from the ISP class were slightly smaller in both subsp. fastidiosa and multiplex (Supplementary Figure S9). Core genes from the Poorly characterized (P) category were removed from the visualizations. Genic GC content and the log10 transformation of r/m were significantly correlated for individual gene alignments of subsp. multiplex (r(3869) = −0.1516, P = 2.2 × 10−16) and subsp. fastidiosa (r(3723) = −0.1396, P = 2.2 × 10−16), but not on subsp. pauca (r(1930) = 0.043, P = 0.0588). However, both significant correlations were small.
Table 6.
χ2 analysis showing statistically significant differences between the number of recombinant and non-recombinant genes from different COG functions within each X. fastidiosa subspecies
| Subspecies | Functional group | Recombinant sum | Non-recombinant sum |
|---|---|---|---|
| Xylella fastidiosa subsp. fastidiosa | Cellular Processes and Signaling (CPS) | 4,721 | 9,469 |
| Information Storage and Processing (ISP) | 6,253 | 9,539 | |
| Metabolism (M) | 7,299 | 22,822 | |
| Multiple categories (MU) | 507 | 2,182 | |
| Poorly characterized (P) | 21,554 | 34,491 | |
| χ2 = 2277.5, df = 4, P < 2.2 × 10−16* | |||
| Xylella fastidiosa subsp. multiplex | Cellular Processes and Signaling (CPS) | 188 | 3,427 |
| Information Storage and Processing (ISP) | 211 | 4,221 | |
| Metabolism (M) | 335 | 6,972 | |
| Multiple categories (MU) | 38 | 777 | |
| Poorly characterized (P) | 625 | 13,559 | |
| χ2 = 4.4525, df = 4, P = 0.3482 | |||
| Xylella fastidiosa subsp. pauca | Cellular Processes and Signaling (CPS) | 4,721 | 9,469 |
| Information Storage and Processing (ISP) | 6,253 | 9,539 | |
| Metabolism (M) | 7,299 | 22,822 | |
| Multiple categories (MU) | 507 | 2,182 | |
| Poorly characterized (P) | 21,554 | 34,491 | |
| χ2 = 2277.5, df = 4, P < 2.2 × 10−16* | |||
Recombinant sum, sum of recombinant genes from all isolates within each subspecies; Non-recombinant sum, sum of non-recombinant genes from all isolates within each subspecies. Bold values correspond to p-values < 0.05. These values have also been marked with asterisk (*).
Statistically significant differences.
Discussion
Compared with its closest relative, Xanthomonas spp. (∼5 Mb and 65% GC), X. fastidiosa (∼2.5 Mb and ∼51% GC) has undergone significant genomic changes. As in other obligate symbionts (Moran et al. 2008; McCutcheon and Moran 2012; Wolf and Koonin 2013), the switch in lifestyle in X. fastidiosa was accompanied by a genome reduction. Compositional changes associated with genome reduction are linked to loss of DNA repair genes, which limits the effects of mutational bias toward C/G to A/T transitions (Moran et al. 2008; Hershberg and Petrov 2010). This was not the case here, as genes associated with DNA repair functions (i.e. MutS, RadA, RecN, RecF, RecO, and AlkA) were present in Xanthomonas spp. and the core/soft-core of X. fastidiosa. Repair genes were hypothetically duplicated in the Xanthomonas-Xylella ancestor and later lost in X. fastidiosa (Martins-Pinheiro et al. 2004). This finding, in addition to the higher GC content observed in Xanthomonas spp., would suggest that the absence of repair gene duplicates in X. fastidiosa facilitated the drop in genome-wide GC content. Previous estimates of the mutation rate of X. fastidiosa [7.6 × 10−7 mutations per site per year; (Vanhove et al. 2019)] are larger than those reported for X. citri [8.4 × 10−8 substitutions per site per year; (Richard et al. 2020)] and, though not directly comparable, smaller than those reported for X. oryzae pv. oryzae [2 × 10−5 mutations per gene per year; (Midha et al. 2017)]. So, the loss of specific gene repair paralogs in X. fastidiosa might have facilitated a species-wide drop in GC content, while the remaining genes aid in maintaining its current nucleotide composition. Yet, not all repair genes are expected to affect GC content (i.e. genes involved in base excision repair correct C/G to A/T mutations, while mismatch repair genes do not) (Garcia-Gonzalez et al. 2012). Future studies directly assessing the mutation rate in X. fastidiosa should be conducted to determine the fidelity of the remaining repair genes.
Another explanation for the drop in GC content observed in X. fastidiosa is environmentally imposed nutritional limitations. Shifts to lower GC content are linked to nutrient-limiting environments (Mann and Chen 2010). During energetic constrains, proteins coded using more energetically costly G/C nucleotides are at a selective disadvantage compared with those favoring A/T nucleotides (Rocha and Danchin 2002). This, in addition to an A/T mutational bias, would result in a GC content drop (Mann and Chen 2010). For X. fastidiosa, xylem-sap, the primary nutrient source in insect mouthparts and in planta, is nutrient limited (Bové and Garnier 2003). In certain xylem-feeding insects, these limitations led to long-term mutualistic associations with Candidatus Baumannia cicadellinicola and Candidatus Sulcia muelleri, which are housed in bacteriocytes (Braendle et al. 2003; Bennett et al. 2014); both C. Baumannia cicadellinicola (∼20.3% GC) and C. Sulcia muelleri (∼22.7% GC) have low GC genomes.
Alternatively, genome reduction is linked to selection-driven gene-loss on accessory genes (Lee and Marx 2012). Recent studies have found that nucleotide composition is more constrained in genes from the core genome compared with the accessory genome (Bohlin et al. 2017, 2018). This pattern was also observed here. So, compositional changes in X. fastidiosa might be the result of selection-driven gene loss eliminating GC-rich accessory genes. This hypothesis should be tested in studies encompassing a larger range of plant-associated bacteria.
Another point to consider is the unique mutation-selection balance on X. fastidiosa. Natural selection can counteract the A/T mutation bias observed in microbial organism (Bohlin et al. 2018). In instances where natural selection is limited, genic nucleotide composition would lean toward lower GC values. In X. fastidiosa, this could be observed in newly introduced clonal populations. Subsp. fastidiosa was introduced to the USA and subsp. pauca was introduced to Italy via a single event ∼150 years ago (Vanhove et al. 2019)) and ∼17 years ago (Giampetruzzi et al. 2017; Vanhove et al. 2019), respectively. Yet, genic GC content was only slightly lower in introduced (52.16% GC in Italy and 51.85% GC in the USA) vs native populations (52.97% GC in Brazil and 51.91% GC in Costa Rica). Very few studies have assessed transitions and transversion rates in X. fastidiosa, with most of them using limited genomic data (Doddapaneni et al. 2006). In those using larger datasets, point mutations consistently contribute less to genome diversity than homologous recombination (Scally et al. 2005; Rogers and Stenger 2012; Vanhove et al. 2019, 2020). While it is unlikely that mutational biases are different in this pathogen compared with others; it is unknown if point mutations in X. fastidiosa are limited, if they have been under detected (i.e. clonal populations), or if they are masked by other evolutionary forces (i.e. recombination). Future studies should leverage the increasing publicly available genome data for this pathogen to explicitly address this issue.
Nucleotide composition is variable within the accessory genome
GC content was more constrained in the core than the accessory genome. Among accessory genes, the cloud genome (<15% strains) could represent either ancestral gene loss in certain lineages or recent gene gain (Davison 1999; Mira et al. 2001). The latter might be more likely since our samples are biased toward newly introduced clonal populations. On the other hand, the soft-core genome (95–99% strains) likely represents recent gene loss events or annotation/assembly errors. Nucleotide composition changes adaptive to a xylem environment (i.e. lower GC) would be observed by identifying compositional differences between these groups. Such a trend is observed here.
Strand-biased nucleotide composition but no strand-biased gene distribution in X. fastidiosa
GC content was lower in X. fastidiosa’s leading strands. This could be explained by differences in the energetic requirements for de novo synthesis of nucleotides and amino acids resulting in strand-biased nucleotide composition (SNC) (Chen and Zhang 2013; Gao et al. 2017). In energy-limiting environments and GC-low genomes, SNC favors the lowest energetic cost for protein-coding genes (Wu et al. 2012). In most bacteria, mutational bias in the lagging strand favors energetically cheaper nucleotides but more expensive protein products (Rocha 2008; Gao et al. 2017). As a result, GC-rich protein-coding genes are preferentially found in the leading strand, particularly if they are highly expressed (Gao et al. 2017). Alternatively, replication-transcription conflicts also influence differences in the nucleotide composition of DNA strands. During replication of the lagging strand, RNA and DNA polymerases produce mutations caused by head-on collisions that are more deleterious than the co-directional mutations on the leading strand (Merrikh et al. 2012). If AT-biased substitutions are more deleterious in the lagging strand, then the lower GC content in the leading strand of X. fastidiosa might be due to the accumulation of A/T mutations otherwise removed from the lagging strand. Nonetheless, the exact mechanism by which replication-transcription conflicts influence genome evolution is still debated. Increase mutagenesis in the lagging strand can be adaptive (Paul et al. 2013; Merrikh and Merrikh 2018), or a product of deleterious mutation/purifying selection balance (Chen and Zhang 2013). On the other hand, non-synonymous substitutions are under strong purifying selection in the leading strand (Schroeder et al. 2020). Which trend better fits X. fastidiosa remains to be determined.
Xylella fastidiosa is one of a few bacteria with no significant GC skew (Gregory and DeSalle 2005). However, GC skew analyses have been conducted using few strains, and therefore, subtle differences might not have been detected. Here, we found that the average number of genes in the lagging versus leading strands was 1055 vs 1030 for subsp. fastidiosa, 1096 vs 1095 for subsp. pauca, and 1127 vs 1079 for subsp. multiplex. Contrary to our observation, estimations based on the predicted location of the origin and terminus of replication projected that 59–60% of X. fastidiosa genes would be located on the leading strand (Retchless et al. 2014). These estimates were obtained using five finished and three draft quality X. fastidiosa genome sequences. Therefore, trends specific to X. fastidiosa might have not been observed. Also, nucleotide compositional changes driven by pressures for rapid and efficient gene expression might not be predominant in slow-growing bacteria such as X. fastidiosa (Bustamante Smolka et al. 2003). This would result in a similar number of genes in either DNA strand as seen here. Bacterial chromosomes lacking the DNA polymerase III alpha subunit polC (such as X. fastidiosa) also have less significant strand-biased gene distribution (Wu et al. 2012). Finally, G/T and A/T nucleotide combinations were predominant in the leading strand, while A/C and G/C combinations were predominant in the lagging strand. This suggests that, in X. fastidiosa, T bases have strong dominance in the leading strand, followed by G bases. This matches other non-polC genomes (Gao et al. 2017) as well as predictions based on mutational and selective pressures (Zhang and Gao 2017).
Gene function, size, and location have an interlinked effect on X. fastidiosa GC content
Genes from the ISP functional class, particularly in the subsp. fastidiosa and multiplex core genomes, were shorter and had lower GC content compared with other genes. The differences were largely the result of a clustered group of ribosomal protein-coding genes. After their removal, both CPS and ISP genes had similar GC content distribution. In X. fastidiosa, survival is tightly linked to efficient replication and movement within xylem vessels (Sicard et al. 2018). Genes involved in “Translation” and “Replication” often had lower GC content than other bacterial genes. Codons that facilitate mRNA folding into a more unstable secondary structure are thought to enable efficient translation initiation (Gu et al. 2010; Bentele et al. 2013). Also, repetitive AT segments and genes with lower GC content found near the replication origin (oriC), facilitate the opening of the DNA double helix and the initiation of replication (Rajewska et al. 2012; Li et al. 2014). Functions associated with “Cell cycle control” and “Signal transduction,” both involved in sensing and responding to external signals (Skerker et al. 2005), also had lower GC content. Our results suggest that nucleotide compositions facilitating replication and growth are favored.
The clustered organization of ribosomal protein genes (ribosomal superoperons) matches that previously described in other bacteria. Such an organization facilitates control during transcription and translation (Lecompte et al. 2002; Fox et al. 2009; Yutin et al. 2012). In fast-growing bacteria, ribosomal protein genes are located near the oriC, as a mechanism to secure more ribosomal proteins, facilitate ribosome assembly, and enable translation (Soler-Bistué et al. 2015). Though ribosomal proteins were clustered in X. fastidiosa, they were not closely associated with the estimated start of the replication in complete genome assemblies fitting expectations for slow-growing bacteria. The ribosomal supercluster was closer to the oriC in subsp. fastidiosa (∼536,660–549,051) and subsp. multiplex (∼547,934–561,894 bp), compared with subsp. pauca (∼1,109,361–1,121,748 bp). Whether this indicates adaptive changes facilitating rapid replication in some X. fastidiosa subspecies or is the result of architectural chromosome changes (though the cluster is outside three major inversion events) remains to be evaluated.
Outside X. fastidiosa, ISP-linked differences in GC content were only found in X. oryzae pv. oryzae and X. oryzae pv. oryzicola. Moreover, ISP genes had lower marginal-GC content in X. oryzae pv. oryzae than in X. oryzae pv. oryzicola. The mechanisms to invade plant tissues are different in each pathovar (i.e. vascular tissue vs plant parenchyma) (Niño-Liu et al. 2006). Gene gain/loss events have mediated the transition between vascular and non-vascular pathovars (Gluck-Thaler et al. 2020). Therefore, other genomic variables (i.e. nucleotide composition, genome architecture, or gene duplication) could also be associated with the mechanisms of plant infection. Xylem-limited pathogens move through long distances in a nutrient-limited environment and cause systemic infection while non-vascular pathogens remain restricted to infection sites formed by living parenchyma cells (Mensi et al. 2014). In this regard, the nutritional limitations imposed by the host environment would be different between pathovars, which could lead to variations in genomic GC content. Another point to consider is that different ISP genes might have distinct evolutionary origins due to genome rearrangement and duplication events. Further analyses focused on Xanthomonas spp. should be conducted to address this question.
Natural selection was associated with gene-specific GC content only in subsp. pauca
Purifying selection was predominant in X. fastidiosa and dN/dS values were similar regardless of the number of ortholog sequences within gene alignments. Previous reports (Bohlin et al. 2017) have found that purifying selection limits the amount of viable variation in genes that are essential for survival, particularly within the core genome. In accordance, our results show that most non-synonymous changes are deleterious. Xylella fastidiosa is thought to have undergone genome reduction (Simpson 2000; Rodriguez-R et al. 2012) and developed a complete, but minimalist, metabolic network (Kurokawa et al. 2016; Gerlin et al. 2020). This and its recent association with multiple crops (Rapicavoli et al. 2018; Sicard et al. 2018) could have resulted in limitations to X. fastidiosa genetic variation. Only few groups of ortholog genes showed signs of positive selection (dN/dS > 1). Most of them belonged to the M class. A less conservative analysis (e.g. a branch-site selection test) might highlight a different pattern, yet, these results indicate that metabolic adaptation is occurring in X. fastidiosa.
Genes favoring higher dN/dS had lower GC content in subsp. pauca. However, we could not establish if GC content was differently favored between non-synonymous vs synonymous changes from our results. Within bacteria, natural selection favors increased synonymous GC content (Hildebrand et al. 2010; Raghavan et al. 2012); so, we expect that a similar trend would be observed in X. fastidiosa. The negative relation between GC content and dN/dS values observed in subsp. pauca could suggest a decrease in GC content favored following a drop associated, perhaps, with its genome reduction. Yet, this correlation (r(1930) = −0.0558, P = 0.0142) was small and future studies should determine its biological relevance, as well as address the potential role of gene expression and transcription in GC content. In addition, it should be noted that in the case of subsp. pauca and subsp. fastidiosa; a significant proportion of our data originates from recently introduced populations. This could limit the action of natural selection due to a recent founder effect.
Intra-subspecific recombination has a variable effect in X. fastidiosa GC content
Recombination occurs between sympatric subsp. pauca strains (Almeida et al. 2008; Coletta-Filho et al. 2017; Francisco et al. 2017), has a phylogenetic and geographic component (Nunney et al. 2013, 2014a, 2014b; Kandel et al. 2017; Landa et al. 2020,), and plays a role in speciation (,Nunney et al. 2014a,) and host switching (,Nunney et al. 2014b; Coletta-Filho et al. 2017; Vanhove et al. 2019,). Yet, our results indicate that intra-subspecific recombination does not have a distinct effect on nucleotide composition. Recombinant core genes had lower GC content than non-recombinant core genes, though this was not observed when genes were subdivided according to function. This suggests that the differences in nucleotide composition among functional groups are not caused by recombination, at least in the core genome. In addition, while r/m rates were significantly correlated with GC content in subsp. multiplex (r(3869) = −0.1516, P = 2.2 × 10−16) and subsp. fastidiosa (r(3723) = −0.1396, P = 2.2 × 10−16) the relationships were small. Whether these trends are biologically meaningful or not, they indicate that homologous recombination might play a role in mediating the nucleotide composition of the accessory genome.
Previous studies have found that GC-biased gene conversion (gBGC) results in increased GC content in recombinant genes (Lassalle et al. 2015). Alternatively, GC content decreases within recombinant regions of highly recombinant genomes (Suerbaum et al. 1998; Falush et al. 2001; Lassalle et al. 2015). A study evaluating 54 bacterial genomes with diverse lifestyles (i.e. endosymbionts and intracellular pathogens, opportunistic pathogens, commensal and free-living bacteria, and obligate pathogens), found that nucleotide composition did not differ between recombinant and non-recombinant regions (González-Torres et al. 2019). While this study focused largely on bacterial species of medical interest, the three plant pathogens included (X. campestris, X. oryzae, and X. fastidiosa) did not show any differentiating trends. The analysis conducted here confirms these results in X. fastidiosa’s core genome but does not eliminate the possibility nucleotide composition in the accessory genome might be linked to recombination.
Conclusion
Xylella fastidiosa has undergone significant biological, ecological, and genomic changes. Thus, it can be a valuable organism to better understand nucleotide compositional changes in bacterial plant symbionts. Our results indicate that GC content has dropped in X. fastidiosa compared with its closest relative. Several hypotheses are presented to explain this drop and should be tested further. Yet, particular focus should be dedicated to better understanding the mutation rate of X. fastidiosa. For this pathogen species, changes in nucleotide composition do exist but are small. It is notable that nucleotide composition of most of the genome of X. fastidiosa is conserved regardless of numerous variables having a statistical effect on it. We hypothesize that distinct evolutionary forces and energetic constraints both drive and limit these small variations. For example, recombination in the core genome and purifying selection would limit the number of nucleotide changes; while recombination in the accessory genome, nutritional limitations in the environment, and mutation/selection biases drive genic GC content drops. Taken together, we show that even in a GC-balanced genome like that of X. fastidiosa, nucleotide changes are observed, and the action of evolutionary forces can be detected.
Acknowledgments
Genome sequencing was performed at the UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory.
Funding
This work has received funding from the PD/GWSS Research Program, California Department of Food and Agriculture, and the European Union’s Horizon 2020 research and innovation program “Xylella fastidiosa Active Containment Through a multidisciplinary-Oriented Research Strategy XF-ACTORS” under grant agreement N. 727987. UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory is supported by NIH instrumentation grant (S10 OD018174).
Conflicts of interest: The authors declare no conflict of interest.
Literature cited
- Almeida RPP. 2018. Emerging plant disease epidemics: biological research is key but not enough. PLoS Biol. 16:e2007020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RPP, Nascimento FE, Chau J, Prado SS, Tsai CW, et al. 2008. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl Environ Microbiol. 74:3690–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almpanis A, Swain M, Gatherer D, McEwan N.. 2018. Correlation between bacterial G+C content, genome size and the G+C content of associated plasmids and bacteriophages. Microb Genom. 4:0–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, et al. 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 1:543–556. [DOI] [PubMed] [Google Scholar]
- Andrews S, Wingett SW, 2018. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 7:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt PF, Hwa T, Petrov DA.. 2005. Substantial regional variation in substitution rates in the human genome: Importance of GC content, gene density, and telomere-specific effects. J Mol Evol. 60:1–6. [DOI] [PubMed] [Google Scholar]
- Aslam S, Lan XR, Zhang BW, Chen ZL, Wang L, et al. 2019. Aerobic prokaryotes do not have higher GC contents than anaerobic prokaryotes, but obligate aerobic prokaryotes have. BMC Evol Biol. 19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi KJ, Rocha EPC, Feil EJ.. 2009. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol. 26:345–355. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA.. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. MBio. 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Blüthgen N.. 2013. Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol. 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J, Eldholm V, Brynildsrud O, Petterson JHO, Alfsnes K.. 2018. Modeling of the GC content of the substituted bases in bacterial core genomes. BMC Genomics. 19:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J, Eldholm V, Pettersson JHO, Brynildsrud O, Snipen L.. 2017. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genomics. 18:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, et al. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 7:169–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botzman M, Margalit H.. 2011. Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 12:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové J, Garnier M.. 2003. Phloem-and xylem-restricted plant pathogenic bacteria. Plant Sci. 164:423–438. [Google Scholar]
- Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, et al. 2003. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 1:e21–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L. 2014. The GC content of bacterial genomes. J Phylogenetics Evol Biol. 2:1–3. [Google Scholar]
- Bustamante Smolka M, Martins D, Winck FV, Santoro CE, Castellari RR, et al. 2003. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics. 3:224–237. [DOI] [PubMed] [Google Scholar]
- Castillo AI, Chacón-Díaz C, Rodríguez-Murillo N, Coletta HD, Almeida RPP.. 2020. Impacts of local population history and ecology on the evolution of a globally dispersed pathogen. BMC Genomics. 21:1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AI, Nelson ADL, Lyons E.. 2019a. Tail wags the dog? Functional gene classes driving genome-wide GC content in Plasmodium spp. Genome Biol Evol. 11:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AI, Tuan S-J, Retchless AC, Hu F-T, Chang H-Y, et al. 2019b. Draft whole-genome sequences of Xylella fastidiosa subsp. fastidiosa strains TPD3 and TPD4, isolated from grapevines in Hou-li, Taiwan. Microbiol Resour Announc. 8:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-H, Lu G, Bork P, Hu S, Lercher MJ.. 2016. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat Commun. 11334:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang J.. 2013. Why are genes encoded on the lagging strand of the bacterial genome? Genome Biol Evol. 5:2436–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta-Filho HD, Francisco CS, Lopes JRS, Muller C, Almeida RPP.. 2017. Homologous recombination and Xylella fastidiosa host–pathogen associations in South America. Phytopathology. 107:305–312. [DOI] [PubMed] [Google Scholar]
- Contreras-Moreira B, Vinuesa P.. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 79:7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornara D, Cavalieri V, Dongiovanni C, Altamura G, Palmisano F, et al. 2017. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. J Appl Entomol. 141:80–87. [Google Scholar]
- Davison J. 1999. Genetic exchange between bacteria in the environment. Plasmid. 42:73–91. [DOI] [PubMed] [Google Scholar]
- Didelot X, Wilson DJ.. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon MM, Sung W, Lynch M, Cooper VS.. 2015. The rate and molecular spectrum of spontaneous mutations in the GC-rich multichromosome genome of Burkholderia cenocepacia. Genetics. 200:935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddapaneni H, Yao J, Lin H, Walker MA, Civerolo EL.. 2006. Analysis of the genome-wide variations among multiple strains of the zplant pathogenic bacterium Xylella fastidiosa. BMC Genomics. 7:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du MZ, Zhang C, Wang H, Liu S, Wei W, et al. 2018. The GC content as a main factor shaping the amino acid usage during bacterial evolution process. Front Microbiol. 9:2948–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C, Paul S.. 2012. Microbial lifestyle and genome signatures. Curr Genomics. 13:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA). 2018. Update of the Xylella spp. host plant database. EFSA J. 16:1–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes AM, Hearn DJ, Agrawal S, Pierson EA, Dunning Hotopp JC.. 2018. Comparative genomics of the Erwinia and Enterobacter olive fly endosymbionts. Sci Rep. 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M.. 2016. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Kraft C, Taylor NS, Correa P, Fox JG, et al. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci U S A. 98:15056–15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerstner KU, von Mering C, Hooper SD, Bork P.. 2005. Environments shape the nucleotide composition of genomes. EMBO Rep. 6:1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE, Wang J, Dasgupta I.. 2009. Many nonuniversal archaeal ribosomal proteins are found in conserved gene clusters. Archaea. 2:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco CS, Ceresini PC, Almeida RPP, Coletta-Filho HD.. 2017. Spatial genetic structure of coffee-associated Xylella fastidiosa populations indicates that cross infection does not occur with sympatric citrus orchards. Phytopathology. 107:395–402. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L.. 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genet Soc Am. 159:907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Lu G, Lercher MJ, Chen WH.. 2017. Selection for energy efficiency drives strand-biased gene distribution in prokaryotes. Sci Rep. 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez A, Rivera-Rivera RJ, Massey SE.. 2012. The presence of the DNA repair genes mutM, mutY, mutL, and mutS is related to proteome size in bacterial genomes. Front. Genet. 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlin L, Cottret L, Cesbron S, Taghouti G, Jacques M-A, et al. 2020. Genome-scale investigation of the metabolic determinants generating bacterial fastidious growth. Am Soc Microbiol. 5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampetruzzi A, Saponari M, Loconsole G, Boscia D, Savino VN, et al. 2017. Genome-wide analysis provides evidence on the genetic relatedness of the emergent Xylella fastidiosa genotype in Italy to isolates from Central America. Phytopathology. 107:816–827. [DOI] [PubMed] [Google Scholar]
- Gluck-Thaler E, Cerutti A, Perez-Quintero AL, Butchacas J, Roman-Reyna V, et al. 2020. Repeated gain and loss of a single gene modulates the evolution of vascular plant pathogen lifestyles. Sci Adv. 6:eabc4516–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Torres P, Rodríguez-Mateos F, Antón J, Gabaldón T.. 2019. Impact of homologous recombination on the evolution of prokaryotic core genomes. MBio. 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TRT, DeSalle R.. 2005. Comparative genomics in prokaryotes. In T. Ryan Gregory, editors. The Evolution of the Genome. Chapter 10. Academic Press. p. 585–675. [Google Scholar]
- Gu W, Zhou T, Wilke CO.. 2010. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA.. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6:e1001115–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A.. 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6:e1001107–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DS. 2002. The diversity of plant pathogens and conservation: Bacteria and fungi. In: K Sivasithamparama, KW Dixon, RL Barrett, editors. Microorganisms in Plant Conservation and Biodiversity. Chapter 9. Dordrecht: Springer. p. 241–267. [Google Scholar]
- Kandel PP, Almeida RPP, Cobine PA, De La Fuente L.. 2017. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol Plant Microbe Interact. 30:589–600. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 21:676–679. [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Seno S, Matsuda H, Ying BW.. 2016. Correlation between genome reduction and bacterial growth. DNA Res. 23:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa BB, Castillo AI, Giampetruzzi A, Kahn A, Román-Écija M, et al. 2020. Emergence of a plant pathogen in Europe associated with multiple intercontinental introductions. Appl Environ Microbiol. 86:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Périan S, Bataillon T, Nesme X, Duret L, et al. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 11:e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O.. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Marx CJ.. 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8:e1002651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J. et al. , 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Zhong ZJ, Zhu PP, Deng EZ, Ding H, et al. 2014. Sequence analysis of origins of replication in the Saccharomyces cerevisiae genomes. Front Microbiol. 5:574–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhou J, Wu Y, Yang S, Tian D.. 2015. GC-content of synonymous codons profoundly influences amino acid usage. G3 (Bethesda). 5:2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR, Sueoka N.. 2002. Asymmetric directional mutation pressures in bacteria. Genome Biol. 3:RESEARCH0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Thompson LR, Stingl U, Hughes AL.. 2015. Selection maintains low genomic GC content in marine SAR11 lineages. Mol Biol Evol. 32:2738–2748. [DOI] [PubMed] [Google Scholar]
- Mann S, Chen YPP.. 2010. Bacterial genomic G+C composition-eliciting environmental adaptation. Genomics. 95:7–15. [DOI] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, et al. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 13:614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17:5–7. [Google Scholar]
- Martins-Pinheiro M, Galhardo RS, Lage C, Lima-Bessa KM, Aires KA, et al. 2004. Different patterns of evolution for duplicated DNA repair genes in bacteria of the Xanthomonadales group. BMC Evol Biol. 4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA.. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- Mensi I, Vernerey MS, Gargani D, Nicole M, Rott P.. 2014. Breaking dogmas: the plant vascular pathogen Xanthomonas albilineans is able to invade non-vascular tissues despite its reduced genome. Open Biol. 4:130116–130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D.. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 4:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh CN, Merrikh H.. 2018. Gene inversion potentiates bacterial evolvability and virulence. Nat Commun. 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Zhang Y, Grossman AD, Wang JD.. 2012. Replication-transcription conflicts in bacteria. Nat Rev Microbiol. 10:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha S, Bansal K, Kumar S, Girija AM, Mishra D, et al. 2017. Population genomic insights into variation and evolution of Xanthomonas oryzae pv. oryzae. Sci Rep. 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA.. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42:165–190. [DOI] [PubMed] [Google Scholar]
- Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, et al. 2017. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol. 34:1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugal CF, Arndt PF, Holm L, Ellegren H.. 2015. Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes. G3 (Bethesda). 5:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, et al. 2015. Gene-wide identification of episodic selection. Mol Biol Evol. 32:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño-Liu DO, Ronald PC, Bogdanove AJ.. 2006. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 7:303–324. [DOI] [PubMed] [Google Scholar]
- Nunney L, Hopkins DL, Morano LD, Russell SE, Stouthamer R.. 2014a. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl Environ Microbiol. 80:1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R.. 2014b. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol. 80:3025–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L, Vickerman DB, Bromley RE, Russell SA, Hartman JR, et al. 2013. Recent evolutionary radiation and host plant specialization in the Xylella fastidiosa subspecies native to the United States. Appl Environ Microbiol. 79:2189–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, et al. 2013. Assembly single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 20:714–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall LM, Rebek EJ.. 2017. Insect vectors and current management strategies for diseases caused by Xylella fastidiosa in the Southern United States. J Integr Pest Manag. 8:1–12. [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 31:3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H.. 2013. Accelerated gene evolution via replication-transcription conflicts. Nature. 495:512–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbu U, Ramos-Onsins SE, Lercher MJ.. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31:1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Kelkar YD, Ochman H.. 2012. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci U S A. 109:14504–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewska M, Wegrzyn K, Konieczny I.. 2012. AT-rich region and repeated sequences—the essential elements of replication origins of bacterial replicons. FEMS Microbiol Rev. 36:408–434. [DOI] [PubMed] [Google Scholar]
- Ramazzotti M, Cimaglia F, Gallo A, Ranaldi F, Surico G, et al. 2018. Insights on a founder effect: the case of Xylella fastidiosa in the Salento area of Apulia, Italy. Phytopathol Mediterr. 57:8–25. [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F.. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35:2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli J, Ingel B, Blanco-Ulate B, Cantu D, Roper C.. 2018. Xylella fastidiosa: an examination of a re-emerging plant pathogen. Mol Plant Pathol. 19:786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless AC, Labroussaa F, Shapiro L, Stenger DC, Lindow SE.. 2014. Genomics of plant-associated bacteria. In: DC Gross, L-P Ann, K Chittaranjan, editors. Genomic Insights into Xylella fastidiosa Interactions with Plant and Insect Hosts. Berlin, Heidelberg: Springer-Verlag. p. 177–202. [Google Scholar]
- Richard D, Pruvost O, Balloux F, Boyer C, Rieux A, et al. 2020. Time-calibrated genomic evolution of a monomorphic bacterium during its establishment as an endemic crop pathogen. Mol Ecol. Doi: 10.1111/mec.15770. [DOI] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, et al. 2009. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 25:2071–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. 2008. The organization of the bacterial genome. Annu Rev Genet. 42:211–233. [DOI] [PubMed] [Google Scholar]
- Rocha C, Danchin A.. 2002. Base composition bias might result from competition for metabolic resources. TRENDS Genet. 18:291–294. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Grajales A, Arrieta-Ortiz ML, Salazar C, Restrepo S, et al. 2012. Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 12:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Stenger DC.. 2012. A conjugative 38 kB plasmid is present in multiple subspecies of Xylella fastidiosa. PLoS One. 7:e52131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Roux C.. 2017. Analytical biases associated with GC-content in molecular evolution. Front Genet. 8:16–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally M, Schuenzel EL, Stouthamer R, Nunney L.. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl Environ Microbiol. 71:8491–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JW, Sankar TS, Wang JD, Simmons LA.. 2020. The roles of replication-transcription conflict in mutagenesis and evolution of genome organization. PLoS Genet. 16:e1008987–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenzel EL, Scally M, Stouthamer R, Nunney L.. 2005. A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl Environ Microbiol. 71:3832–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- Sicard A, Zeilinger AR, Vanhove M, Schartel TE, Beal DJ, et al. 2018. Xylella fastidiosa: insights into an emerging plant pathogen. Annu Rev Phytopathol. 56:181–202. [DOI] [PubMed] [Google Scholar]
- Simpson AJG. 2000. The complete genome sequence of the plant pathogen Xylella fastidiosa. Biochem Soc Trans. 28:A102–A102. [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT.. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334–e1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Leitch IJ, Mucina L, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc Natl Acad Sci U S A. 111:E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Bistué A, Mondotte JA, Bland MJ, Val ME, Saleh MC, et al. 2015. Genomic location of the major ribosomal protein gene locus determines Vibrio cholerae global growth and infectivity. PLoS Genet. 11:e1005156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, et al. 1998. Free recombination within Helicobacter pylori. Proc Natl Acad Sci U S A. 95:12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV.. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme F, Koebnik R, Bekel T, Berger C, Boch J, et al. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium. J Bacteriol. 187:7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaondo Z, Molina L, Segura A, Duque E, Ramos JL.. 2016. Analysis of the core genome and pangenome of Pseudomonas putida. Environ Microbiol. 18:3268–3283. [DOI] [PubMed] [Google Scholar]
- Vanhove M, Retchless AC, Sicard A, Rieux A, Coletta-Filho HD, et al. 2019. Genomic diversity and recombination among Xylella fastidiosa subspecies. Appl. Environ. Microbiol. 85:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove M, Sicard A, Ezennia J, Leviten N, Almeida RPP.. 2020. Population structure and adaptation of a bacterial pathogen in California grapevines. Environ Microbiol. 22:2625–2638. [DOI] [PubMed] [Google Scholar]
- Vicente JG, Holub EB.. 2013. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol Plant Pathol. 14:2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV.. 2013. Genome reduction as the dominant mode of evolution. Bioessays. 35:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Qu H, Wan N, Zhang Z, Hu S, et al. 2012. Strand-biased gene distribution in bacteria is related to both horizontal gene transfer and strand-biased nucleotide composition. Genom Proteom Bioinf. 10:186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI.. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 7:e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gao F.. 2017. Quantitative analysis of correlation between AT and GC biases among bacterial genomes. PLoS One. 12:e0171408–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HQ, Ning LW, Zhang HX, Guo FB.. 2014. Analysis of the relationship between genomic GC content and patterns of base usage, codon usage and amino acid usage in prokaryotes: similar GC content adopts similar compositional frequencies regardless of the phylogenetic lineages. PLoS One. 9:e107319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequence files are available upon request. Published sequence data are available at GenBank; the accession numbers are listed in Supplementary Table S1. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14067449.