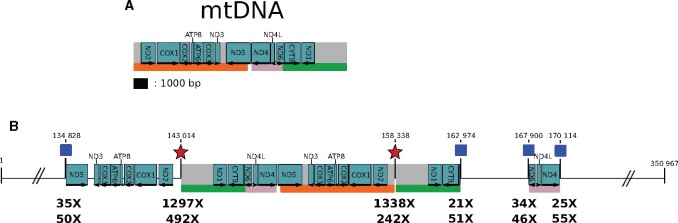
Figure 2.
A large NUMT in the S. nonagrioides genome. (A) Map of the mitogenome assembly. Protein-coding genes are represented by blue rectangles, in which arrows show their directions. The names of the genes are indicated in the blue rectangles or on top of them. The scale at the bottom of the mtDNA applies also for (B). (B). Map of scaffold scf7180000018078_1 showing the coordinates of the four blastn hits obtained using the mitogenome as a query on the Sesamia nonagrioides nuclear assembly. Only the NUMT sequence is drawn to scale, the rest of the scaffold is not, as indicated by broken lines. The scale of the NUMT is the same as in (A). The four hits are located between each coordinate and the orange, green and pink rectangles help to visualize which parts of the mtDNA have been integrated and possibly rearranged in the nuclear DNA. The nuclear-mitochondrial junctions (blue squares) are supported by short reads (depth of the trimmed reads indicated by the top numbers under the junctions). Long reads support both nuclear-mitochondrial junctions (depth of the reads longer than 7 kb indicated by the bottom numbers under the junctions) and mitochondrial breakpoints (red stars). Indeed, we visually checked that some long reads straddle on the mitochondrial breakpoints and the nuclear genome. The coverage at the mitochondrial breakpoints is much higher because these breakpoints are also mapped by all reads originating from the cytoplasmic mitogenomes. The first, second and fourth nuclear–mitochondrial junctions are also supported by PCR.