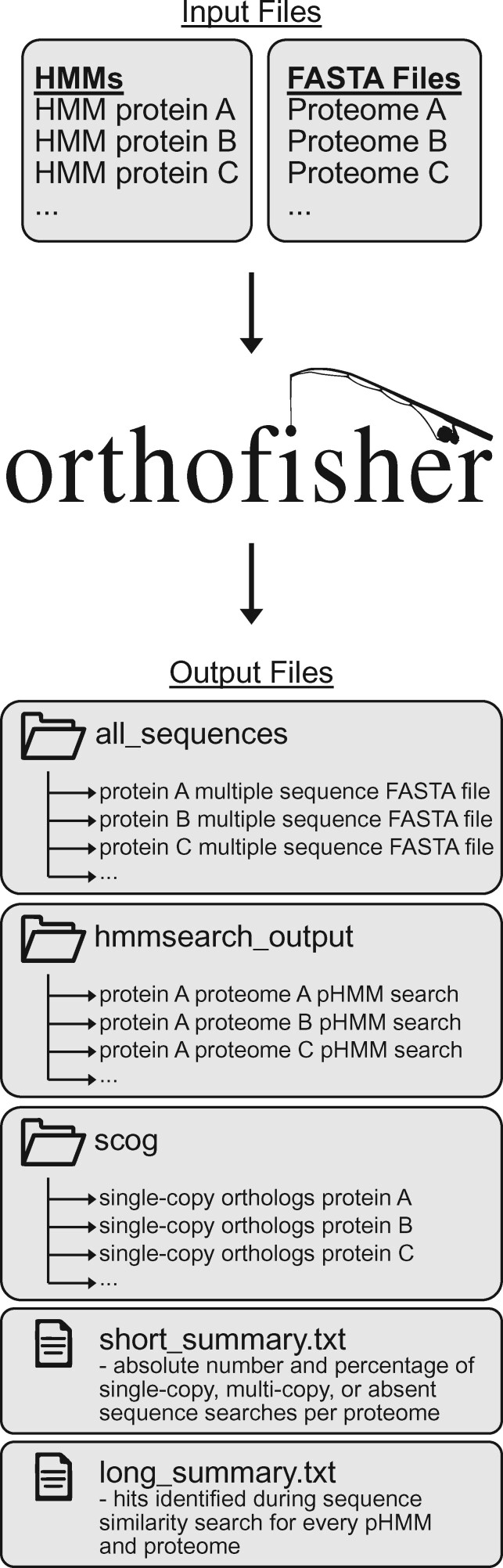
Figure 1.
Workflow overview for orthofisher. orthofisher takes two files as input, which specify the location of query pHMMs and the FASTA files wherein sequence similarity searches will be performed. orthofisher then outputs three directories and two text files that summarize results and facilitate downstream analyses.