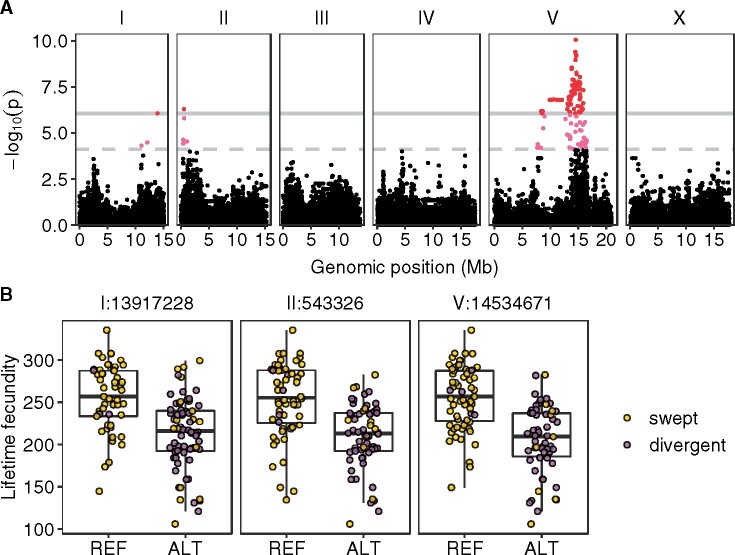
Figure 3.
Three QTL were identified in GWA mapping of lifetime fecundity variation in 121 C. elegans wild strains. (A) Manhattan plot indicating GWA mapping results. Each point represents an SNV that is plotted with its genomic position (x-axis) against its −log10(p) value (y-axis) in mapping. SNVs that pass the genome-wide EIGEN threshold (the dotted gray horizontal line) and the genome-wide BF threshold (the solid gray horizontal line) are colored pink and red, respectively. (B) Tukey box plots showing lifetime fecundity between strains with different genotypes at the peak marker position in each QTL. Each point corresponds to a C. elegans strain and is colored gold for swept strains and purple for divergent strains. On the x-axis, REF represents strains with the N2 reference allele and ALT represents strains with the alternative allele.