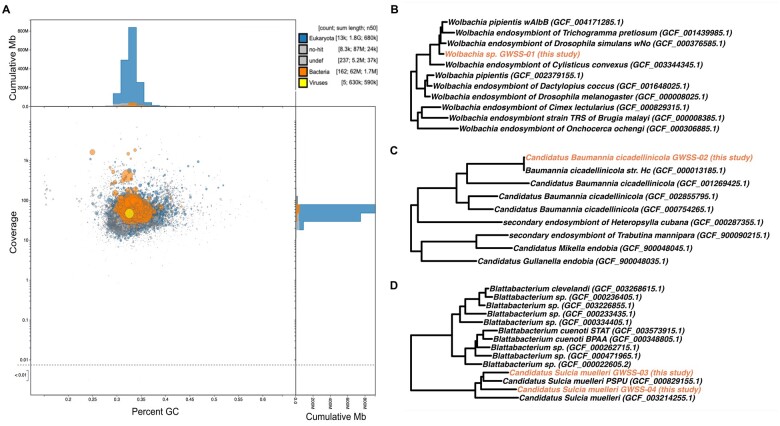
Figure 2.
Endosymbiont assessment in genome and identification. (A) BlobTools2 visualization of H. vitripennis scaffolds showing taxa-colored GC coverage plot. Each circle represents a scaffold in the assembly, scaled by length, and colored by superkingdom (eukaryota = blue, bacteria = orange, viruses = yellow, and unidentified = gray). On the x-axis is the average GC content of each scaffold and on the y-axis is the average coverage of each scaffold to the draft assembly. The marginal histograms show cumulative genome length (Mb) for coverage (y-axis) and GC content bins (x-axis). (B) Placement of Wolbachia sp. GWSS-01 (colored in orange) in the GTDB phylogenetic tree. (C) Placement of Ca. Baumannia cicadellinicola GWSS-02 (colored in orange) in the GTDB phylogenetic tree. (D) Placement of Ca. Sulcia muelleri GWSS-03 and GWSS-04 (colored in orange) in the GTDB phylogenetic tree.