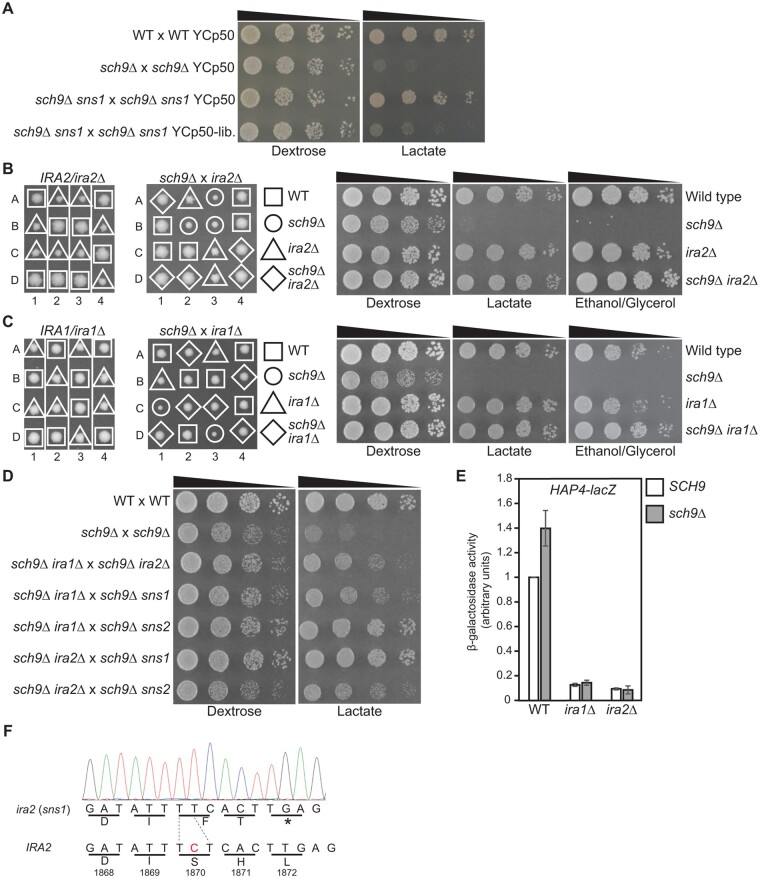
Figure 5.
SNS1 and SNS2 are IRA2 and IRA1, respectively. (A) A sns1 mutation is complemented by a genomic DNA library plasmid. Wild type (BY4741 × BY4742), sch9Δ × sch9Δ (PPY745), and sch9Δ sns1 × sch9Δ sns1 (PPY741) mutant strains were transformed with control vector YCp50 or the sns1-complementing library plasmid, YCp50-lib. (pPP322). Transformants were serially diluted and spotted on YNBcasD (Dextrose) and YNBcasL (Lactate) plates. (B) Tetrad analysis of IRA2/ira2Δ (ZLY6578, left panel) and ira2Δ (ZLY6601) × sch9Δ (ZLY6649) (middle panel) on YPD plates. Wild type (BY4741) and isogenic sch9Δ (ZLY6649), ira2Δ (ZLY6609), and sch9Δ ira2Δ (ZLY6661) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates (right panel). (C) Tetrad analysis of IRA1/ira1Δ (ZLY6656, left panel) and sch9Δ (ZLY6649) × ira1Δ (ZLY6648) (middle panel) on YPD plates. Wild type (BY4741) and isogenic sch9Δ (ZLY6650), ira1Δ (ZLY6648), and sch9Δ ira1Δ (PPY754) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates (right panel). (D) Diploid strains obtained from the crossings among sch9Δ ira1Δ (PPY755), sch9Δ ira2Δ (ZLY6661), sch9Δ sns1 (PPY733, PPY734), and sch9Δ sns2 (ZLY6619, ZLY6626) mutant strains as indicated were serially diluted and spotted on YPD and YPL plates. BY4741 × BY4742 and sch9Δ × sch9Δ (PPY745) strains were included as controls. (E) β-galactosidase activities of a HAP4-lacZ reporter gene in wild type (BY4741), ira1Δ (ZLY6647), and ira2Δ (ZLY6609) mutant strains with or without an sch9Δ deletion mutation grown in YNBcas5D medium. The HAP4-lacZ reporter gene activity in the wild type strain is set as 1. (F) IRA2 from sns1 mutant cells has a frameshift mutation. The chromatogram shows the sequence of IRA2 containing the frameshift mutation from a sns1 mutant. The nucleotide C missing in the IRA2 gene from the sns1 mutant was highlighted in red in the wild type IRA2 sequence.