Figure 2.
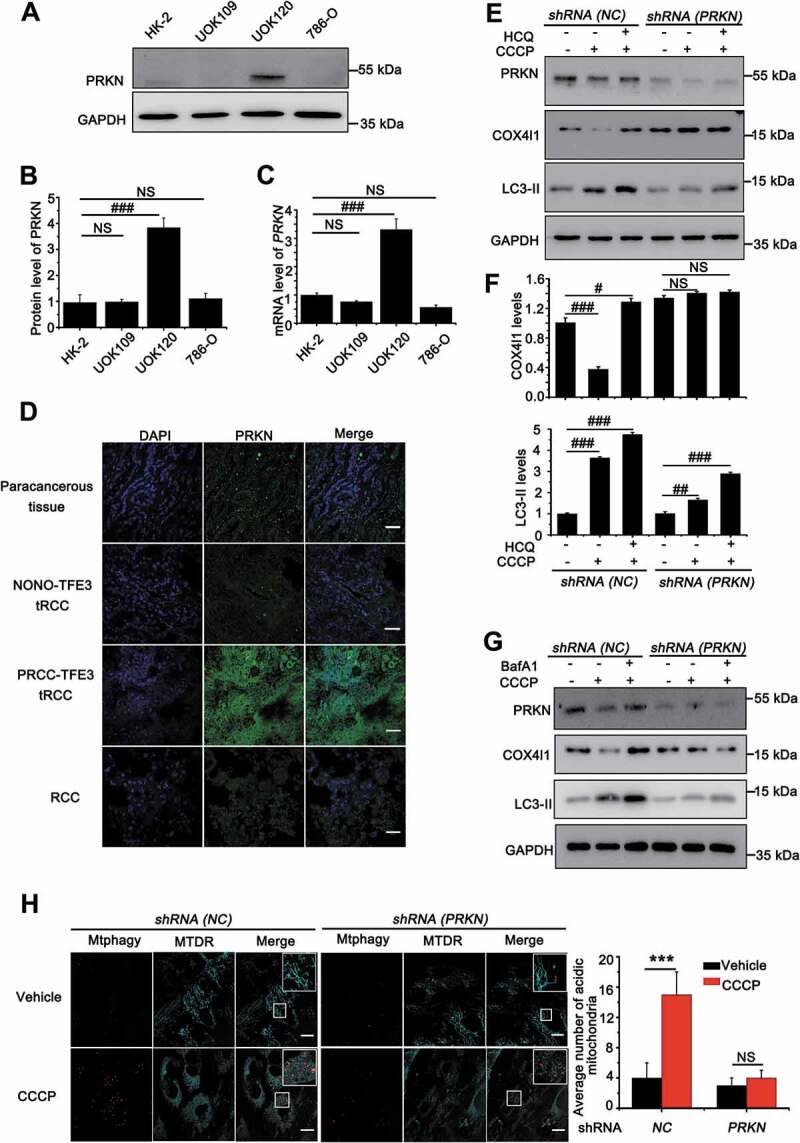
Amplification of PRKN boosts mitophagy in UOK120 cells. (A) Representative blots of PRKN in HK-2, UOK109, UOK120 and 786-O cells. (B) Densitometric analysis of PRKN:GAPDH in (A). Data are presented as mean ± s.e.m from 3 independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 (One-way ANOVA with Dunnett posttests). (C) Analysis of the mRNA levels of PRKN by real-time PCR in HK-2, UOK109, UOK120 and 786-O cells. Data are presented as mean ± s.e.m from 3 independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 (One-way ANOVA with Dunnett posttests). (D) Renal paracancerous tissues, Xp11.2 tRCC, or RCC tissue were sectioned and stained with a fluorescent anti-PRKN antibody (green) and counter stained with DAPI to reveal nuclei, followed by immunofluorescence photomicrographic analysis. Scale bars: 30 μm. (E) UOK120 cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (PRKN), and then PRKN, LC3-II and COX4I1 were examined by western blot after treatment with CCCP (10 μM) for 6 h in the presence or absence of HCQ (30 μM). (F) Densitometric analysis of COX4I1:GAPDH (upper) and LC3-II:GAPDH (down). Data are presented as mean ± s.e.m from 3 independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 (One-way ANOVA with Dunnett posttests). (G) UOK120 cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (PRKN), and then PRKN, LC3-II and COX4I1 were examined by western blot after treatment with CCCP (10 μM) for 6 h in the presence or absence of Baf A1 (200 nM). (H) UOK120 cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (PRKN) followed by staining with the Mtphagy dye and MitoTracker Deep Red, and then visualization of Mtphagy dye and MitoTracker Deep Red using live-cell imaging microscopy after treatment with CCCP (10 μM) for 12 h. Scale bars: 20 μm. Right panel, quantification of acidic (red-only) mitochondria in different group, data are presented as mean ± s.e.m from 3 independent experiments (10 cells/group). GAPDH was used as a loading control. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed t-test). NS, no significance
