Figure 5.
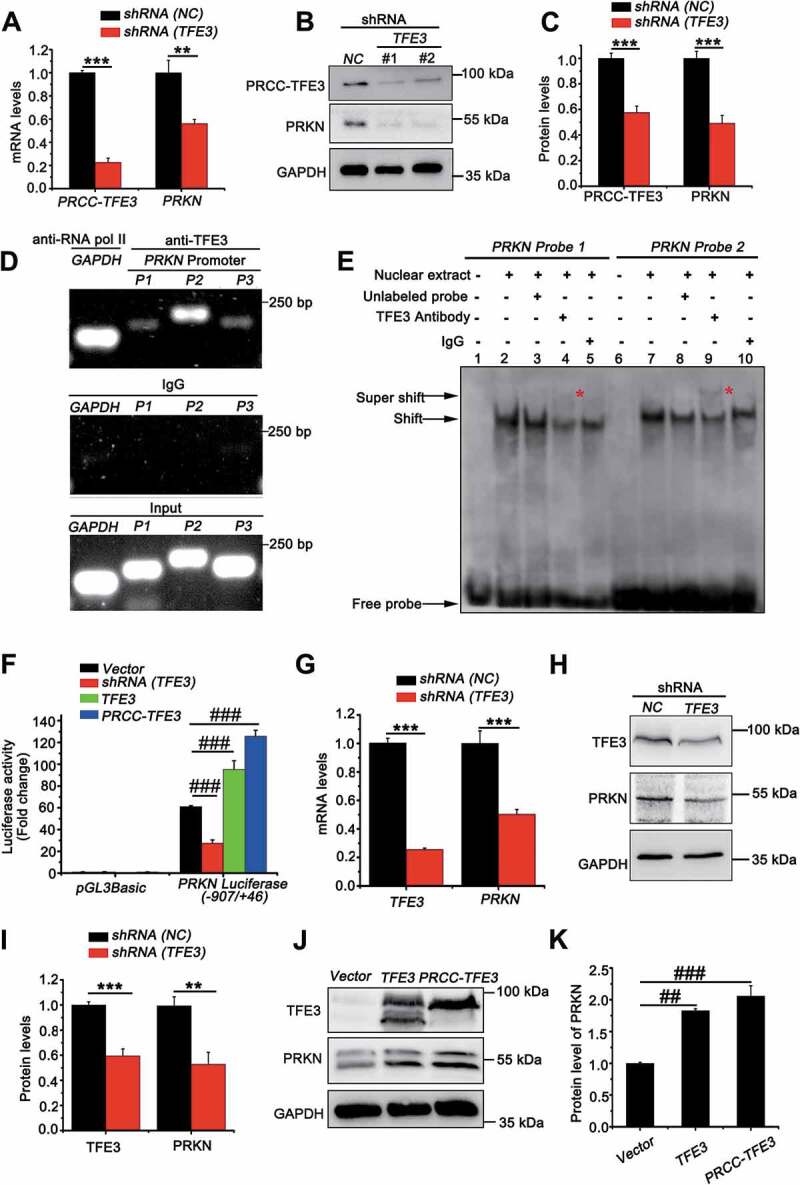
PRKN is the target gene of TFE3 and PRCC-TFE3. (A) UOK120 cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (TFE3), followed by analysis of the mRNA levels of PRKN and PRCC-TFE3 by real-time PCR. Data are presented as mean ± s.e.m from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed t-test). (B) UOK120 cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (TFE3) and PRKN and PRCC-TFE3 were examined by western blot. (C) Densitometric analysis of PRCC-TFE3:GAPDH and PRKN:GAPDH. Data are presented as mean ± s.e.m from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed t-test). (D) HEK293T cells were analyzed by the ChIP assay using an anti-TFE3 antibody and immunoprecipitated DNA fragments were amplified by PCR. P1, PRKN promoter region (−109/-8); P2, PRKN promoter region (−397/-388) and P3, PRKN promoter region (−673/-657). (E) TFE3 binding was evaluated by EMSA using nuclear extract from HEK293T cells and two different biotin-labeled double-stranded oligonucleotide probes (probe 1 and 2) containing TFE3 putative binding sites. The unlabeled specific TFE3 competitor oligonucleotides (100-fold molar excess) and TFE3 antibody are indicated above each lane. DNA-protein complexes are indicated by arrows in each panel. Supershifted bands are indicated by red asterisks. (F) HEK293T cells were transiently co-transfected with an empty vector (vector), or vectors expressing shRNA (TFE3), TFE3 or PRCC-TFE3, together with the human PRKN promoter region (−907/+46) constructs or PGL3 Basic, followed by a luciferase reporter gene assay. The pRL-TK vector was co-transfected to normalize transfection efficiencies. Results are presented as a luciferase/Renilla ratio. Data are presented as mean ± s.e.m from 3 independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 (One-way ANOVA with Dunnett posttests). (G) HEK293T cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (TFE3), followed by analysis of the mRNA levels of PRKN and TFE3 by real-time PCR. Data are presented as mean ± s.e.m from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed t-test). (H) HEK293T cells were infected with Lenti.shRNA (NC) or Lenti.shRNA (TFE3) and PRKN and TFE3 were examined by western blot. (I) Densitometric analysis of TFE3:GAPDH and PRKN:GAPDH. Data are presented as mean ± s.e.m from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed t-test). (J) HEK293T cells were transiently transfected with empty vector (vector), or vectors expressing TFE3, or PRCC-TFE3 and PRKN and TFE3 were examined by western blot. (K) Densitometric analysis of PRKN:GAPDH. GAPDH was used as a loading control. Data are presented as mean ± s.e.m from 3 independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 (One-way ANOVA with Dunnett posttests)
