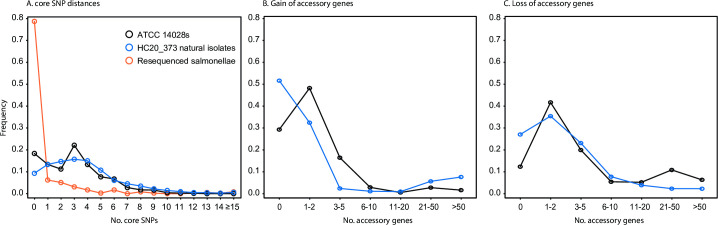
Fig 1. Core and accessory genetic diversity within 496 HC20_373 genomes from strains stored in laboratory collections or isolated from nature.
A). Core SNP distances. Numbers of non-repetitive SNPs in an all against all comparison of pairs of genomes within each category. Maximum numbers of core SNP differences: Natural isolates, 30; ATCC 14028s, 16; resequenced genomes, 16. B). Numbers of additional accessory genes according to a pan-genome of all isolates in an all against all comparison of pairs of genomes within each category. C). Numbers of deleted accessory genes according to a pan-genome of all isolates in an all against all comparison of pairs of genomes within each category. ATCC 14028s (black): 172 strains from laboratory sources or from laboratory infections. HC20_373 natural isolates (blue): 324 strains from all other sources. Resequenced salmonellae (red; only in part A): 285 pairs of genomes that were sequenced twice for the UCCUoW 10K genomes project [14]. Additional details on the accessory genes in parts B and C can be found in S2 Fig.