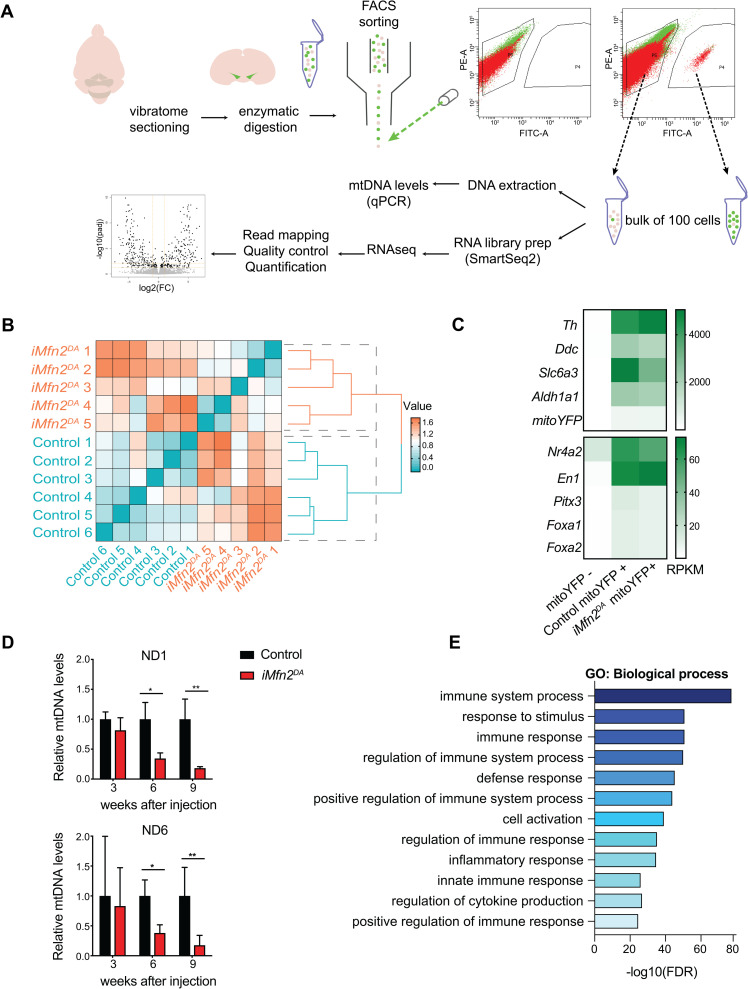
Fig 4. Immune response and mtDNA depletion in tamoxifen-injected iMfn2DA mouse brains.
(A) Experimental workflow: vibratome sections were enzymatically digested and mechanically triturated resulting in a single cell suspension; the bulk of mitoYFP positive (mitoYFP+) cells and mitoYFP negative (mitoYFP-) cells were collected by FACS; samples were used for either mtDNA quantification or for RNA library preparation followed by RNAseq. (B) Hierarchical clustering analysis of RNAseq data from DA neurons isolated from n = 5 iMfn2DA and n = 6 control mice. (C) Heatmap showing that RNA levels (Reads Per Kilobase Million, RPKM) of genes encoding DA neuron markers (Th, Ddc, Scl6a3, and Aldh1a1) and transcription factors (Nr4a2, En1, Pitx3, Foxa1 and Foxa2) are highly enriched in mitoYFP+ samples. Data are shown as mean of the RPKM in control (n = 6), iMfn2DA (n = 5), and mitoYFP- (n = 3) mice. (D) Relative mtDNA levels (ND1/18S rRNA and ND6/18S rRNA) measured in FACS-sorted DA neurons. *p< 0.05, **p<0.01, n = 4. Data are shown as mean ± SD. (E) Gene ontology (GO) analysis showing the most dysregulated biological processes in FACS-sorted DA neurons isolated from iMfn2DA mice at 3 weeks after tamoxifen injection.