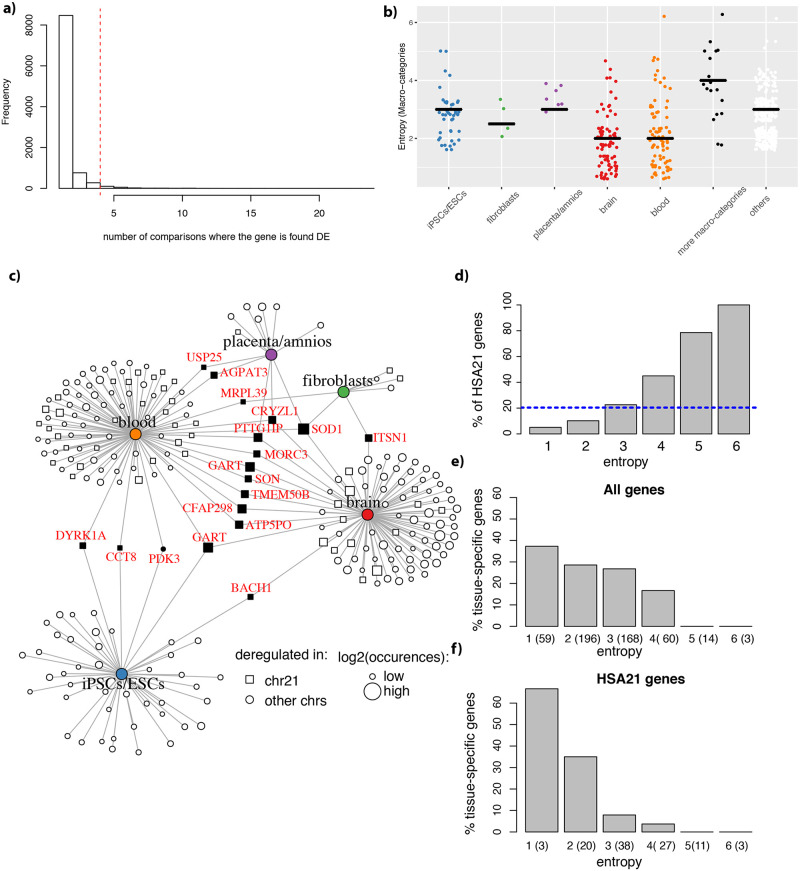
Fig 2. Tissue macro-categories show both HSA21 and non-HSA21 genes that are consistently DE.
a) Histogram of DE genes’ occurrences. Genes in the right 5% are in the right of the red dot line, corresponding to genes found DE in at least 4 comparisons b) Entropy distribution across consistently DE genes. Each dot is a gene, the horizontal bar indicates the median entropy for each group. All genes consistently DE in iPSCs/ESCs were also found DE in at least another macro-category up to 4 other macro categories. Gene consistently DE in fibroblasts were also found DE in one or two macro-categories. Similarly, genes consistently DE in placenta/amnios were found DE also in 2 or 3 other macro categories. For brain and blood tissue we did detect a fraction specifically DE only in one of the two macro-categories, however, also in this case we detected genes DE in more macro-categories (1 to 4 for brain; 1 to 5 for blood). In black are genes that are consistently DE at the tissue level in more than one macro-category. In white, genes that are not consistently DE in any specific macro categories but are consistently DE across tissue. c) Bipartite graph showing the tissue macro-categories where we could find consistently DE genes at the tissue level. For each tissue macro-category, we plotted the frequency of the number of occurrences. The red dotted line indicates the minimal number of occurrences chosen for a gene to be DE in a given tissue macro-category. Genes mapping on HSA21 are represented by squares, otherwise by circles. Node size is proportional to the log2 of the number of occurrences across all comparisons. Of note, most consistently DE genes are HSA21. d) Barplot for the percentage (%) of HSA21 genes for each entropy level. The dotted line indicates the percentage (%) of HSA21 genes in the top-500 DE genes. e) Barplot showing the percentage (%) of tissue specific genes for each entropy level, considering all the top 500 genes f) As in e). but, only considering the top 500 genes mapping on HSA21.