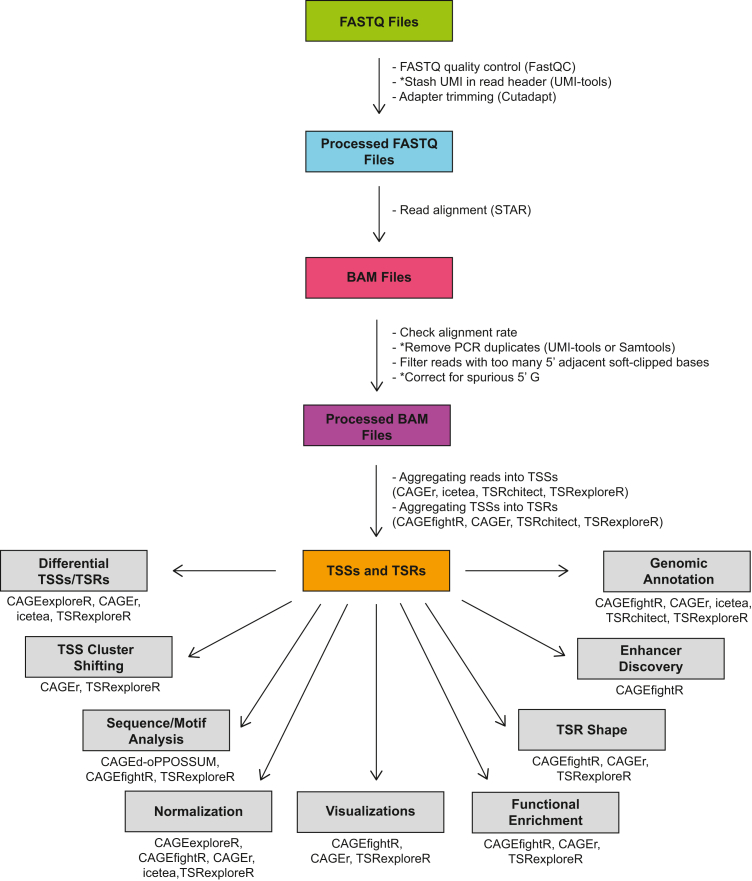
Figure 3.
Computational processing of TSS mapping data
A general workflow for processing and analysis of TSS mapping data is shown, with software that can be used for each step indicated. Asterisks indicate optional steps. More information on each piece of software listed here can be found at the following URLs: FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc); UMI-tools (https://github.com/CGATOxford/UMI-tools); Cutadapt (https://cutadapt.readthedocs.io/en/stable); STAR (https://github.com/alexdobin/STAR); Samtools (http://www.htslib.org); CAGEr (https://www.bioconductor.org/packages/release/bioc/html/CAGEr.html); icetea (https://www.bioconductor.org/packages/release/bioc/html/icetea.html); TSRchitect (https://www.bioconductor.org/packages/release/bioc/html/TSRchitect.html); TSRexploreR (https://zentnerlab.github.io/TSRexploreR/index.html); CAGEexploreR (https://github.com/edimont/CAGExploreR); CAGEd-oPPOSSUM (http://cagedop.cmmt.ubc.ca/CAGEd_oPOSSUM/); CAGEfightR (https://www.bioconductor.org/packages/release/bioc/html/CAGEfightR.html).