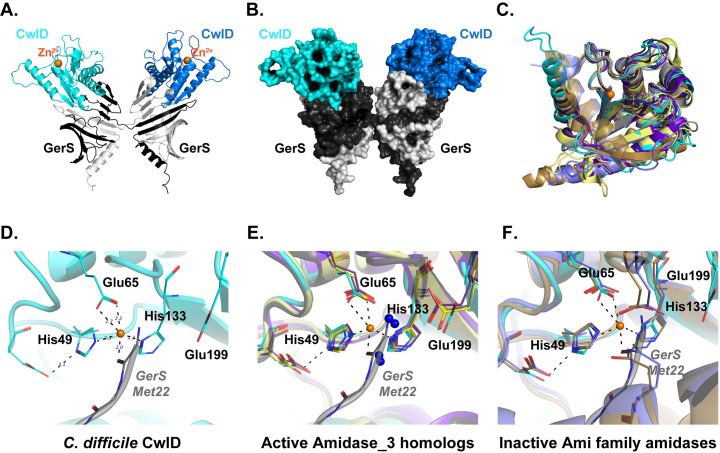
Fig 2. Structure of the CwlD:GerS complex.
(A) Shown is the cartoon representation of the CwlD:GerS complex within the crystal with CwlD in blue tones and GerS in grey. The crystallographic asymmetric unit contains a single protomer of each with the 2 GerS/2 CwlD assembly formed by domain swapping of the GerS monomers. (B) The assembly is shown in space filling representation. (C) Least squares superposition of CwlD (cyan throughout) and its homologs performed using Superpose [44] in CCP4. (D) Close-up view of the metal binding site for CwlD with GerS from a fortuitous crystal packing (grey) providing coordination of the ion. (E) CwlD homolog structures (CwlD Paenibacillus polymyxa PDBID 1JWQ, unpublished; C. difficile CD2761 amidase_3 family member, 4RN7, unpublished; Nostoc punctiforme, AmiC2, 5EMI [29]; and C. difficile Cwp6, 5J72 [52]) in the active form have the Zn2+ atom accessible to the solvent. They also have water molecules serving as 1–2 ligands (blue spheres), one of which is coordinated by a conserved glutamate. (F) CwlD homolog structures (Bartonella henselae AmiB PDBID 3NE8 [22] and E. coli AmiC 4BIN [21]) in the inactive forms have residues from their respective inhibitory helices providing Zn2+ ligands.