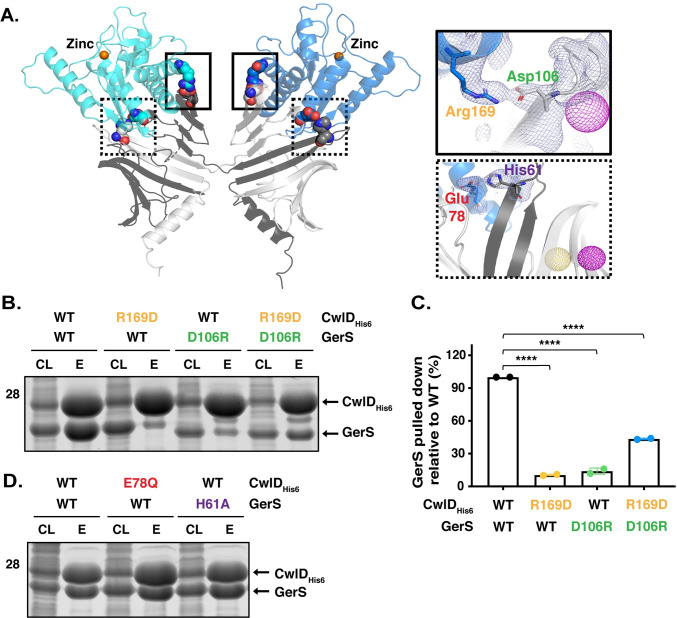
Fig 7. A salt bridge between CwlDR169 and GerSD106 is important for CwlD:GerS binding.
(A) Specific interactions between CwlD and GerS observed in the structure. Salt bridge formation between CwlD Arg169 (R169) and GerS Asp106 (D106) is highlighted in the solid boxes and between CwlD Glu78 (E78) and GerS His61 (H61) in the dotted boxes. The light blue mesh shows the heavy atom phased electron density after density modification and building contoured at 1 sigma. The yellow mesh represents the anomalous signal at 3 sigma for the selenomethionine derivative and the maroon represents the sodium iodide derivative after scaling, outlier rejection and phasing within Autosharp (Global Phasing Limited). (B) Coomassie stain of co-affinity purifications of wild-type or R169D CwlD-His6, with wild-type GerS or the D106R variant. The indicated proteins were produced in E. coli and purified using Ni2+- affinity resin. Cleared lysate (CL) and eluate (E) fractions were analyzed using Coomassie staining. (C) Percentage of GerS pulled down with CwlD relative to the amount detected in the wild-type CwlD:GerS complex, which was set at 100%. The mean of two assays from two independent co-affinity purifications is shown, and dots represent each of those co-affinity purifications. Statistical significance relative to the wild-type complex was determined using a one-way ANOVA and Tukey’s test. **** p < 0.0001. (D) Coomassie stain of cleared lysate and elution fractions from co-affinity purifications of His-tagged CwlD:GerS, CwlDE78Q:GerS, or CwlD:GerSH61A.