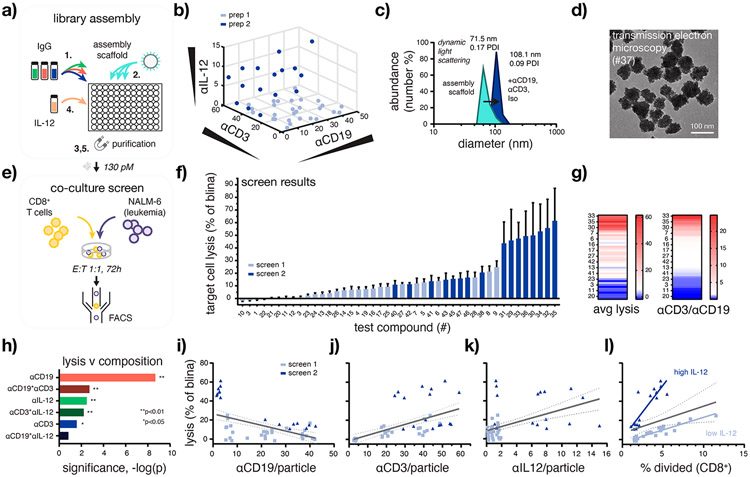
Figure 3.
High-throughput assembly and screening enables rapid identification of BiTEokines that induce efficient leukemia cell lysis. (a) Schematic of test compound library assembly. (b) Structural diversity of the BiTEokine library and (c,d) representative test compound size/morphology as measured by antibody fluorescence intensity, dynamic light scattering, and transmission electron microscopy, respectively. (e) Schematic of coculture assay conditions and (f) parallel screening results rank-ordered by drug-induced lysis of CD19+ NALM-6 leukemia cells by primary human CD8+ T cells. (g) Heatmaps illustrating concordance between target cell lysis the ratio of αCD3 to αCD19 per BiTEokine and (h) significance of antibody components to measured lysis values as modeled by multivariate least-squares regression. Univariate composition-function relationships reporting NALM-6 leukemia cell lysis versus (i–k) antibody abundance per particles and (l) T cell division. Values in (f) represent mean ± SEM of 2–3 T cell donors, each analyzed in duplicate. Lines in (i–l) report linear regressions with 95% confidence intervals. E:T, effector-to-target cell ratio. *p < 0.05, **p < 0.01.