Abstract
Back and neck pain are significant healthcare burdens that are commonly associated with pathologies of the intervertebral disc (IVD). The poor understanding of the cellular heterogeneity within the IVD makes it difficult to develop regenerative IVD therapies. To address this gap, we developed an atlas of bovine (bos taurus) caudal IVDs using single-cell RNA-sequencing (scRNA-seq). Unsupervised clustering resolved 15 unique clusters, which we grouped into the following annotated partitions: nucleus pulposus (NP), outer annulus fibrosus (oAF), inner AF (iAF), notochord, muscle, endothelial and immune cells. Analyzing the pooled gene expression profiles of the NP, oAF and iAF partitions allowed us to identify novel markers for NP (CP, S100B, H2AC18, SNORC, CRELD2, PDIA4, DNAJC3, CHCHD7 and RCN2), oAF (IGFBP6, CTSK, LGALS1 and CCN3) and iAF (MGP, COMP, SPP1, GSN, SOD2, DCN, FN1, TIMP3, WDR73 and GAL) cells. Network analysis on subpopulations of NP and oAF cells determined that clusters NP1, NP2, NP4 and oAF1 displayed gene expression profiles consistent with cell survival, suggesting these clusters may uniquely support viability under the physiological stresses of the IVD. Clusters NP3, NP5, oAF2 and oAF3 expressed various extracellular matrix (ECM)-associated genes, suggesting their role in maintaining IVD structure. Lastly, transcriptional entropy and pseudotime analyses found that clusters NP3 and NP1 had the most stem-like gene expression signatures of the NP partition, implying these clusters may contain IVD-progenitor cells. Overall, results highlight cell type diversity within the IVD, and these novel cell phenotypes may enhance our understanding of IVD development, homeostasis, degeneration and regeneration.
Keywords: Intervertebral Disc, Annulus Fibrosus, Nucleus Pulposus, Single-Cell RNA Sequencing Analysis, Tissue-Specific Progenitor Cells, Bovine
1. Introduction
Low back and neck pain are pressing public health concerns which affect up to 80% of the adult population (1), and domestically cost approximately $134.5 billion (2). The etiology of chronic back pain is multifactorial and complex (3); however, 39-42% of back pain cases involve the intervertebral disc (IVD) (4). This strong association between IVD pathology and back pain highlights the importance of developing regenerative IVD therapies (5, 6). However, advances in regenerative IVD therapies are limited by a poor understanding of IVD biology and lack of consensus regarding the cell types that support IVD health and homeostasis. This information is critical for developing therapies that can stimulate endogenous cell populations to heal the IVD or choosing appropriate cell sources for exogenous cell delivery strategies.
The IVD is a fibrocartilaginous tissue that connects adjacent vertebrae in the spine and permits three-dimensional motions of the spinal column (7). IVDs are composed of two main functional regions: nucleus pulposus (NP) and annulus fibrosus (AF). The NP is the gelatinous core of the IVD that is derived from the notochord (8). The healthy NP contains rounded, chondrocyte-like cells in a randomly-oriented matrix of type II collagen fibers and proteoglycans (9). The AF is a fiber-reinforced, angle-ply laminate, derived from the sclerotome, which condenses in concentric circles around the developing NP (10, 11). The laminated AF structure varies with radial position. The outer AF (oAF) contains elongated, fibroblast-like cells in a matrix of type I collagen, while the inner region of the AF (iAF) contains rounded, fibrocartilage cells in a matrix of type I and II collagens that gradually transitions to the NP region (12). The IVD structure and matrix composition is reasonably well-described and known to relate to function (13, 14), yet little information is known about the heterogeneous populations of cells that make up the NP, iAF and oAF regions (15, 16).
The first studies attempting to understand the unique phenotype of IVD cells used microarrays to compare the gene expression of NP and AF tissues to articular cartilage in rat (17), dog (18) and cow (19) models. These early analyses successfully determined sets of differentially expressed genes (DEGs) that distinguish the IVD from articular cartilage, and the NP from the AF. However, bulk transcriptional assays mask the cellular heterogeneity in the NP and AF regions (20, 21). RNA in situ hybridization studies attempted to transcriptionally profile single cells in the bovine IVD, and successfully identified novel markers of AF and NP cells (22). However, this method relied on known RNA probes and could only assess binary expression. Other studies employed a combination fluorescence-activated cell sorting and gene expression analysis to define IVD cell phenotypes (23), but were similarly limited by requiring known cell markers. Therefore, more robust, unbiased and quantitative methods would be required to find novel markers of heterogeneous IVD subpopulations.
In recent years, single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool to evaluate the gene expression levels of individual cells and to identify heterogeneous subpopulations within a tissue (24, 25). Recently, musculoskeletal researchers have begun to incorporate single-cell omics to investigate rare subpopulations of cells involved in osteoarthritis and cartilage regeneration (26). IVD researchers have begun using sophisticated technologies to develop spatiotemporal proteomic atlases of the human IVD throughout degeneration (27) and recently started using single-cell omic technologies. To date, there have only been three published IVD scRNA-seq papers, which analyze cellular heterogeneity in human (28), rat (29) and cow (30) IVDs. These studies identified novel phenotypic biomarkers of NP and AF cells, and provided some insights into IVD cell heterogeneity. However, a higher resolution investigation of rare subpopulations of IVD cells and of their functional roles in IVD homeostasis is still required to develop our understanding of IVD physiology, pathology and regeneration.
The aims of this study are to assemble a scRNA-seq atlas of the caudal bovine (bos taurus) IVD, predict phenotypic markers of IVD cell populations and use bioinformatic methods to determine functional roles of these cells in IVD homeostasis (Fig. 1). Bovine IVDs are commonly used as a large animal model for IVD research because their NP morphology, mechanical loading, nutrient transport environment and marker genes are concordant with human IVDs (31). We annotate cell type through gene vectors defining each population identified through differential gene expression analysis (DGEA). Cell population annotations are further verified using our custom unsupervised annotation pipeline, which leverages over 6 million scRNA-seq training data points across numerous tissues and cell types. This is followed by network analysis, which uses Gene Set Enrichment Analysis of Gene Ontology (gseGO) and Cnet plots to provide biological interpretations of the variable gene expression data (32), to develop functional interpretations of each subpopulation in IVD homeostasis. Lastly, we used a combination of transcriptional entropy scoring and pseudotime analysis to make predictions about the stem/progenitor characteristics of each subpopulation and resolve differentiation lineages. Collectively, this data allowed us to identify numerous novel markers of NP, oAF and iAF cells, resolve multiple novel cell populations not typically found in IVD, characterize the unique functional roles of heterogeneous subpopulations of NP and oAF cells, and discover potential progenitor cell populations within the IVD.
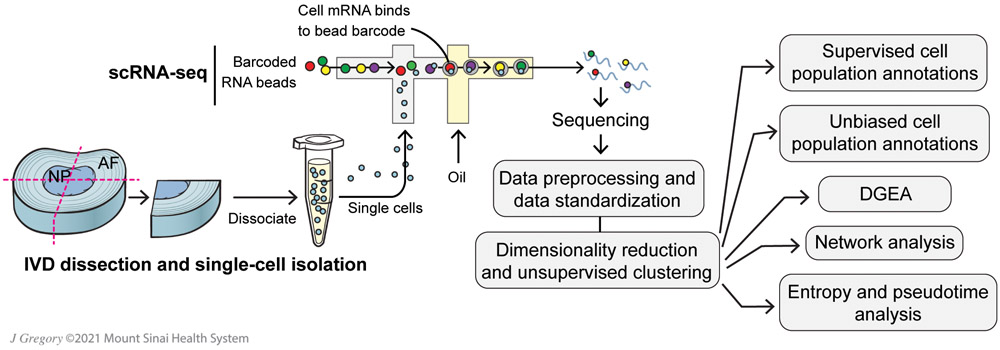
Figure 1: Schematic representation of scRNA-seq analysis pipelines.
Bovine IVDs were quartered, digested and processed for scRNA-seq analysis with numerous downstream bioinformatic analysis pipelines. Differential gene expression analysis (DGEA).
2. Materials and Methods
2.1. IVD Dissection and Single-Cell Isolation
Intervertebral discs (IVDs) were surgically dissected from bovine tails to remove all fat, muscle and ligamentous tissue. Dissections were carefully conducted to ensure little tissue or blood remained on isolated IVDs. IVDs were surgically isolated from the tails of 2 to 3 year-old cows (N=3 animals) purchased from a local abattoir (Springfield Meat Co., Richlandtown, PA). Bovine tails were processed within 4 hrs of slaughter. We isolated two IVDs from two animals and one IVD from the third animal. Samples were labeled by bovine animal (e.g., B6 means bovine number 6) and disc level (e.g., D3 means the third disc from the abattoir cut site). Isolated IVDs were serially washed aseptically with 1X phosphate buffered saline (PBS, Fisher Scientific, Waltham, MA), 70% ethanol (Fisher Scientific), a wash solution of 1.5% Fungizone (Fisher Scientific) and 3% penicillin-streptomycin (PS, Fisher Scientific), then 1X PBS. Cleaned IVDs were cut into quarters and stored overnight at 4°C in MACS Tissue Storage Solution (Miltenyi Biotec, Bergish Gladbach, Germany). After dissection, IVDs were quartered so that each sample contained all IVD regions, from the outermost AF through the innermost NP.
The next day, quartered IVDs were minced into small pieces (~1 mm2), digested using 0.2% pronase (Fisher Scientific) for 1 hr, then digested overnight using 400 U/mL collagenase I (Fisher Scientific). Both digestion solutions were prepared using high-glucose Dulebbco’s Modified Eagle’s Medium (DMEM, Fisher Scientific) supplemented with 2% PS, and tissues were digested in a humidified incubator at 37°C and 20% O2 with agitation. Resulting single-cell suspensions were concentrated to a density of 1x106 cells/mL in Dulbecco's PBS (Sigma Aldrich).
2.2. Single-Cell RNA-Sequencing (scRNA-Seq)
Cells were processed according to Chromium 3’ Gene Expression V3 Kit (10X Genomics, Pleasanton, CA) using manufacturer’s guidelines. An estimated 5,000 cells were loaded onto each channel with a targeted cell recovery of 3,000 cells/sample. This was followed by sequencing on a S1 NovaSeq chip (Illumina Inc., San Diego, CA). Qubit 3 (Fisher Scientific) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) were used for quality check of cDNA. The output BAM file, a binary text file containing the sequence alignment data from sequencing, was processed through 10X Genomics Cell Ranger software v5.0.0. Sequencing generated scRNA-seq data with ~20,000 2x150bp reads per cell and 14,555 total cells sequenced, compiled to over 2B reads with an average 97% mapping rate.
2.3. scRNA-seq Data Processing and Data Standardization
To generate a counts matrix, the sequenced files from each independent sample were processed through 10X Genomics Cell Ranger software v5.0.0. The raw base call files were demultiplexed using Cell Ranger mkfastq pipeline to generate FASTQ files. Cellranger count pipeline was applied to the FASTQs to perform alignment against BosTaurus ARS-UCD 1.2.102 reference build, filtering, barcode counting and Unique Molecular Identifier counting. The feature-barcode matrices were analyzed through a series of open-source R platforms, including Seurat (33), ClusterProfiler (32) and Slingshot (34).
Quality control metrics were applied to the raw gene-barcode dataset (Fig. S2). First, data was filtered to remove genes detected in less than 5 cells per bovine sample. Next, cell barcodes with fewer unique molecular identifiers than the majority of cell barcodes from the sample (i.e., below 1,000) were removed (Figs. S2A & S2D). This processing step removes dead cells and debris infiltration. Next, cell barcodes with a total number of gene counts significantly higher than barcodes within the same sample (i.e., greater than 30,000) were removed (Figs. S2B & S2E). This processing step removes potential doublet cells. Cell barcodes with a high percentage of mitochondrial gene expression were identified through valleys in bimodality of expression. Depending on the sample, this ranged from 5% to 25% of gene detection (Figs S2C & S2F). This processing step removes predicated cells in autophagy. As a representation of this process, Figure S2 visualizes pre- and post-filtered cell populations from sample B7D3 (Fig S2).
Individual sample datasets were linked with the Merge() function followed by recommended standardization steps. The dataset was log normalized with the NormalizeData() Seurat function followed by the ScaleData() Seurat function, where normalization standardizes gene expression between cells to reduce biasing by cell total transcription and scaling data shifts mean expression for every gene across all cells to 0 and standardizes variance of each gene to 1. These reduce processing and technical bias between sample origins and highlighters biologic variability.
2.4. Dimensionality Reduction and Unsupervised Clustering
Principal Component Analysis (PCA) was done for primary dimensionality reduction. Elbow Plot visualization identified 22 dimensions necessary to capture essential data, while also reducing noise effects. Unsupervised graph-based clustering was performed with a low-resolution parameter (0.6). Annotated cluster data is visualized with Uniform Manifold Approximation and Projection (UMAP).
BuildClusterTree() was utilized to inform populational distances between annotated clusters. This function computed distance relationships based on Euclidean distance matrix through average expression of variable genes within a subpopulation.
2.5. Supervised Cell Population Annotations
Differential gene expression analysis (DGEA) was done with Seurat to find top differentially expressed genes (DEGs) for each cluster. We generated top hit differential genes for each cluster with the FindAllMarkers() function. The top genes were identified by log base 2-fold change in expression levels (i.e., avg_logFC) value and difference value. Difference score is calculated as the difference between percent of cells in a cluster expressing a gene and percent of cells outside of a cluster expressing a gene. Expression value represents scaled expression of the specified gene on an individual cell barcode ID. Using a combination of top DEGs and canonical markere gene expression, clusters were grouped into partitions and annotated. Key annotation markers for supervised cell populations are listed in Table 1. Since the iAF is largely understudied, our key annotation markers for this partition were chosen as a combination of NP and oAF markers validated in the literature.
Table 1.
Key annotation markers for supervised cell population annotations.
| Partition | Key Annotation Markers |
|---|---|
| NP (19, 20, 35, 36) | COL2A1, ACAN, HIF1A, SOX9, VCAN |
| oAF (35, 37) | COL1A1, LUM, COL5A1, COL12A1, SFRP2 |
| iAF (22, 35) | COL1A1, FBN1, COL2A1, ACAN, LUM |
| Muscle (38-41) | CNN1, ACTA2, TAGLN, CALD1, SMTN, MYLK |
| Endothelial (42-45) | CDH5, LMO2, PECAM1, KDR, VEGA, FLT1, VWF, CXCR4 |
| Notochord (19, 46) | KRT8, KRT18, KRT19, SHH, TBXT |
| Immune (47-52) | CD68, SRGN, LAPTM5, CD44, IL1RN, IL1B |
2.6. Unbiased Cell Population Annotation Pipeline
Unbiased cluster annotation was performed using a customized, multi-layer dense neural network deconvolution pipeline. The network was trained to deconvolve single-cell expression data into cell phenotype distributions using pseudo-bulk mixtures of 256 unique cell types obtained from publicly available scRNA-seq data. The training data set consisted of over 100 unique altas studies, encompassing 6.2 million single cells. Using this framework, the tool assisted in providing unbiased evidence for cell type annotation by identifying expression patterns shared between cells in our scRNA-seq dataset and the closest cell types found in the training dataset.
2.7. DGEA Pipeline
For partitions with multiple subpopulations of cells, additional DGEA was conducted to determine phenotypic biomarkers of each partition and determine marker genes for subpopulations of cells within each partition. DGEA was done with Seurat, as previously described. To determine gene biomarkers for each cell partition (i.e. NP or oAF), gene expression significance was compared between all cells in the partition with all cells outside that cell partition. By filtering with stringent parameters (avg_logFC > 1.5 and difference > 0.5) for positive gene expression we yield comprehensive gene vectors specific to a cell type. To investigate gene markers for individual clusters, we run a FindAllMarkers() function on subsetted Seurat objects with only the cell types and clusters of interest. This yields a gene vector with highly variable genes between clusters within a partition.
2.8. Network Analysis Pipeline
Individual clusters were compared to populations within their partition using Gene Set Enrichment Analysis of Gene Ontology (gseGO). Gene vectors for a particular subpopulation in comparison to the other clusters are generated using FindAllMarkers(). The input vector for the analysis is subsetted for avg_logFC >0.5. The network analysis liberally filters for pathways with a minimum of 3 genes and up to 2,000 genes annotated within it. Filtered gene vectors are compared to the “org.Bt.eg.dB” bovine reference. The gseGO analysis provides biological interpretations of the variable gene expression data through whole gene vector analysis, providing a robust alternative to manual cell cluster annotation from DGE analysis. Cnet plots were used to display the linkages of genes and biological processes as a network for each NP and oAF cluster (32).
2.9. Entropy and Pseudotime Analysis Pipelines
The mathematical calculation of Shannon entropy as a measurement of stemness used in this paper is based upon the original implementation proposed and detailed extensively by Tessechndorf and Enver (53). In this seminal work, the differentiation potency of a single cell is approximated by computing it’s signaling promiscuity, or entropy; whereby, more stem-like cells have a higher degree of variability in their gene expression when compared to more differentiated cells. Entropy is estimated by calculating the entropy rate of a random-walk markov chain generated using a given protein-protein interaction (PPI) network superimposed with the gene expression of each individual single cell. Entropy calculation was implemented in Tensorflow 2.0, with support for rapid batch processing and GPU/TPU acceleration. Calculation of entropy for the bovine IVD samples was performed using a Google Cloud VM instance consisting of a 4-core Intel Xeon E5-2630 2.3GhZ CPU running on an NVIDIA V100 SMX2 GPU with 25GB RAM using a PPI network previously profiled for use with entropy estimation. Following scoring, spearman correlations were calculated for all genes, comparing log-normalized expression to entropy score with each cell serving as a unique datapoint. Multiple testing correction was performed using bonferroni adjustment. Genes with a positive correlation to entropy are considered overexpressed in high-entropy (stem-like) cell populations, conversely genes with a negative correlation to entropy are considered overexpressed in low-entropy (differentiated) cell populations.
Pseudotime analysis was done with the Slingshot version 2.0 pipeline (34). as.SingleCellExperiment() from the Seurat package converted seurat objects to class ‘SingleCellExperiment’ (SCE). The SCE object is processed through the slingshot() function where a minimum spanning tree (MST) is constructed on cell clusters in order to define a global lineage structure in a reduced dimensionality space (PCA). No supervision for start or end clusters is provided for the analysis. Principal curves are fitted onto the reduced dimension dataset to compute pseudotime scores for each lineage predicting cell level transcriptional states. Ggplot is used to visualize the pseudotime score comparisons between subpopulations.
3. Results
3.1. scRNA-seq Data Reveals Heterogeneous Intervertebral Disc Cell Populations
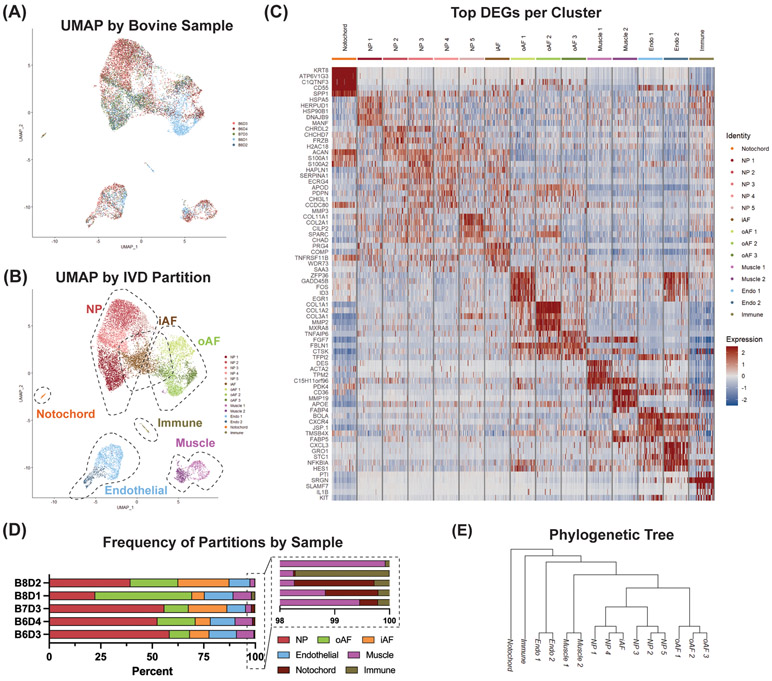
We performed scRNA-seq analysis on 5 intervertebral discs (IVDs) isolated from 3 bovine tails (Fig. 2A). Unsupervised graph-based clustering of 14,555 cells integrated from the 5 samples resolved 15 unique clusters based on nearest neighbor approximations (Fig. 2B). Differential gene expression analysis (DGEA) yielded significant gene expression per cluster (Fig. 2C). The heatmap visualizes the top 5 differentially expressed genes (DEGs) within each cluster sorted by log base 2-fold change (avg_logFC) value. The heatmap subsets each cluster into a randomly generated 60 cell count population for visualization. Using uniform manifold approximation and projection (UMAP) plots, hierarchical clustering and gene expression signatures of published canonical markers (Table 1), we annotated clusters into the following population partitions: nucleus pulposus (NP), outer annulus fibrosus (oAF), inner AF (iAF), notochord, muscle, endothelial and immune (Fig. 2B).
Figure 2: scRNA-seq analysis reveals heterogeneous clusters of IVD cells.
(A) UMAP of cell populations annotated by sample origin. 5 samples comprise the dataset, spanning 3 independent bovine animals. Samples are labeled by animal number and disc level (e.g., B6D3 means bovine animal 6, disc number 3). (B) Partition annotation of identified clusters. (C) Heatmap of top differentially expressed genes (DEGs) per cluster identified by average log base 2-fold change (avg_logFC value). Cell IDs are subsetted for 60 representative cells per cluster. Expression value represents scaled expression of the specified gene on an individual cell barcode ID. (D) Frequency of each IVD partition per bovine sample. (E) Phylogenetic tree outlining population level distance relationships to predict divergence points in global dataset.
We observe all identified partitions across each of the contributing sample IDs in similar proportions (Fig. 2A & 2D). Within individual clusters, there is an observed contribution by each sample origin, except cluster oAF2. oAF2 is composed of 1029 cells and sample B8D1 contributes 1014 (99%) of those cells (Fig. 5B). To investigate potential biasing from B8D1 across the rest of the analysis, we excluded B8D1 and re-clustered our dataset. Re-clustering our dataset without sample B8D1 resulted in the identification of 15 similar clusters (Fig. S1). We observed the loss of cluster oAF2 and the discovery of Endo3. We believe these results are a byproduct of reduced cell counts with a constant clustering resolution. This analysis supports that B8D1 provides the oAF2 population; however, without it we see limited downstream biasing. Importantly, the core partitions (i.e., NP, iAF, oAF and notochord) and their gene expressions in both analyses are similar; thereby providing support for diminished effects of population biasing from sample B8D1.
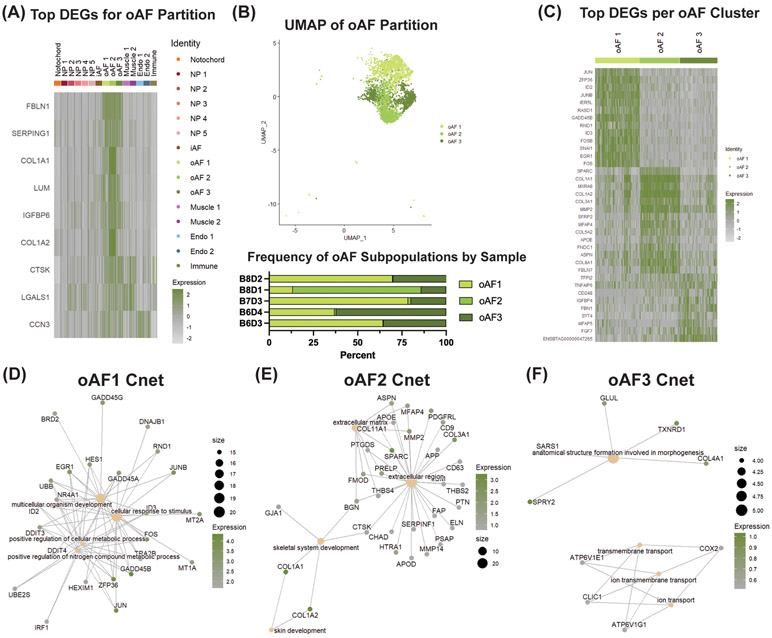
Figure 5: scRNA-seq analysis finds novel oAF cell marker genes, heterogeneous subpopulations of AF cells, and their function.
(A) DGEA between pooled oAF subpopulations and all other clusters. (B) UMAP of oAF partition and frequency of oAF subpopulations per IVD sample. (C) DGEA between oAF subpopulations compared to one another. CNET plots displaying linkages of DEGs and associated biological processes for (D) oAF1, (E) oAF2 and (F) oAF3. Tan dots represent GO terms with size corresponding to the number of genes related to that term. Green/gray dots represent genes with color corresponding to avg_logFC score between the cluster of interest and the other populations for that particular gene. Expression value represents scaled expression of the specified gene on an individual cell barcode ID.
A phylogenetic tree was generated to identify population level organization of the clusters, which further provides population level interpretations of our single-cell dataset. The phylogenetic tree highlights similarity within population partitions, with NP, oAF, endothelial and muscle clusters branching adjacently to other clusters from the same partition (Fig. 2E). Interestingly, despite the iAF population sharing developmental lineage from oAF, the iAF cluster branched adjacent to NP clusters, NP1 and NP4, suggesting more similar gene expression patterns.
3.2. Unbiased Cell Population Annotation Confirms IVD Cell Partitions
To confirm cell population annotations, we used an unbiased cell population annotation pipeline. This analysis overlays our newly generated bovine gene expression data with published scRNA-seq datasets (i.e., training dataset), then annotates cells using the closest cell type within the training dataset. Using our custom unbiased annotation tool we observed high concordance between supervised and unbiased cell population annotations for common cell types (Fig. 3). Cells in the endothelial partition were annotated as endothelial cells with a high score of 0.99. Endothelial cells have been commonly explored in scRNA-seq atlases; thus, the high concordance between our supervised annotation and our unbiased annotation pipeline verifies our cell populations are correctly annotated. We achieved a similar result for our muscle partition; whereby, cells in this partition were labeled as myocytes and myoblasts with scores of up to 0.60 and 0.25, respectively. Cells found in the immune partition were identified as macrophages and hematopoietic stem cells with scores up to 0.25 and 0.25, respectively.
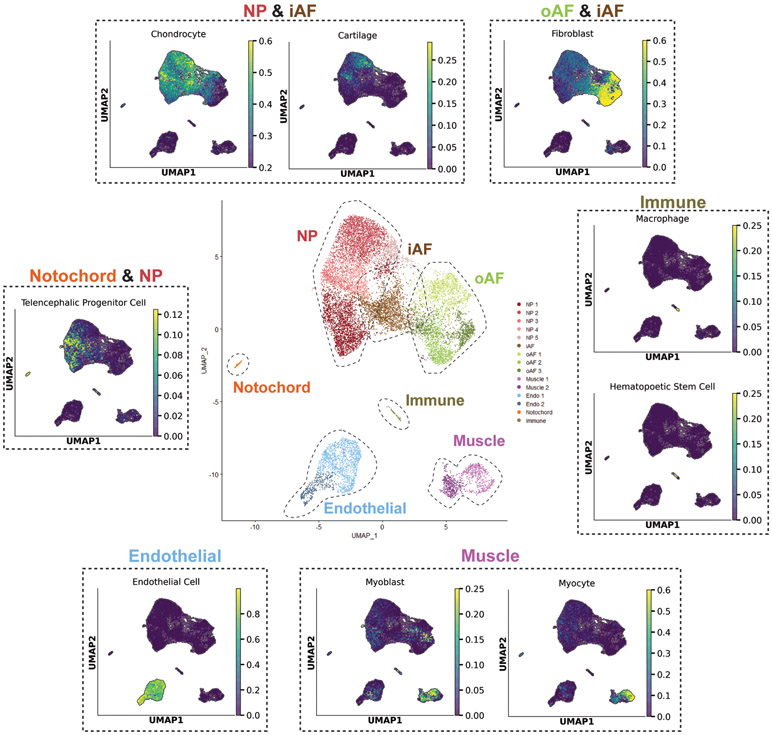
Figure 3: Custom unbiased cell population annotation pipeline corroborates supervised IVD partition annotations.
Center UMAP shows supervised annotations of scRNA-seq dataset. Surrounding UMAPs (dashed lines) were generated by unbiased cell population annotation tool. The unbiased cell population annotation tool annotates cells based on how closely their gene expression matches the gene profiles of annotated cells in published scRNA-seq datasets. UMAPs generated by the tool were organized based on which partition(s) they annotated. For example, many cells in the NP and iAF partitions were categorized as chondrocytes and cartilage using this tool. Scores range from 0 to 1, with higher scores indicating a greater likelihood of correctly annotating cells.
The unbiased cell population annotation tool was capable of making accurate predictions for specific cell types found within the IVD (i.e., NP, notochord, oAF and iAF). scRNA-seq databases for the IVD are extremely rare; thus, the annotation tool could annotate IVD-specific cell types as the closest cell type within the training dataset. Cells in the NP partition were annotated as chondrocytes and cartilage with scorers up to 0.60 and 0.30, respectively. oAF cells were annotated as fibroblasts with scores up to 0.60. These mid-range annotation scores for the NP and oAF partitions highlight that these cells have unique features from chondrocytes and fibroblasts, respectively. Cells in the iAF partition were annotated as both chondrocytes and fibroblasts, demonstrating the fibrocartilaginous nature of the iAF. Interestingly, cells in the notochord partition and the NP3 cluster were annotated as telencephalic progenitor cells with low scores of 0.12. These clusters were found to display progenitor characteristics (Section 2.7), and this unbiased annotation tool further corroborates this finding.
The high concordance of our cluster annotations using supervised and unbiased methods provides confidence that we have correctly annotated all cell clusters. This was an important first step in establishing an atlas of the bovine caudal IVD. Furthemore, it allowed us to continue our analysis of novel cell markers for the NP, notochord oAF and iAF partitions, and make inferences about the functional relevance of subpopulations within these partitions.
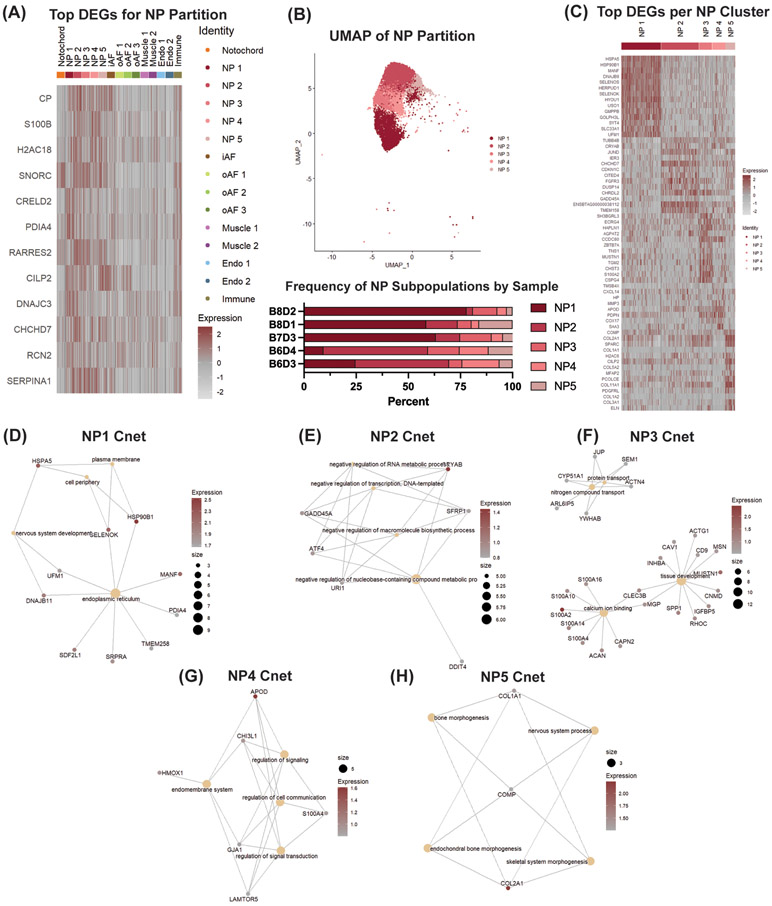
3.3. Analysis of the NP Partition and NP Cell Subpopulations
To better elucidate novel phenotypic biomarkers of NP cells, we compared the pooled gene signature of clusters NP1, NP2, NP3, NP4 and NP5 to the pooled gene signature of all other identified clusters. Through this DGEA, we found that CP, S100B, H2AC18, SNORC, CRELD2, PDIA4, RARRES2, CILIP2, DNAJC3, CHCHD7, RCN2 and SERPINA1 were highly expressed in the NP (Fig 4A). The genes listed in the heatmap generated from this analysis are based on top gene hits by avg_logFC expression values and difference values (Section 4.7). CP, S100B, H2AC18, SNORC, CRELD2, PDIA4, DNAJC3, CHCHD7 and RCN2 have not been previously identified as significant NP markers, highlighting the power of scRNA-seq analysis to elucidate novel phenotypic biomarkers. We observed concordant gene signatures between the NP and notochord partitions for SNORC and RARRES2. While the notochord cluster shared some gene expression patterns with the NP clusters, the notochord cluster was distinguished by an upregulation of KRT8, ATP6V1G3, C1QTNF3, CD55 and SPP1 (Fig. 2C).
Figure 4: scRNA-seq analysis finds novel NP cell marker genes, heterogeneous subpopulations of NP cells and their function.
(A) DGEA between pooled NP subpopulations and all other clusters. (B) UMAP of NP partition and frequency of NP subpopulations per IVD sample. (C) DGEA between NP subpopulations compared to one another. CNET plots displaying linkages of DEGs and associated biological processes for (D) NP1, (E) NP2, (F) NP3, (G) NP4 and (H) NP5. Tan dots represent GO terms with size corresponding to the number of genes related to that term. Red/gray dots represent genes with color corresponding to the expression value between the cluster of interest and the other populations for that particular gene. Expression value represents scaled expression of the specified gene on an individual cell barcode ID.
We next independently analyzed clusters NP1, NP2, NP3, NP4 and NP5 to characterize their gene signatures and elucidate the potential roles each of these populations have in IVD homeostasis (Fig. 4B). Top DEGs, listed with average avg_logFC in each cluster, were identified (Fig. 4C), then Cnet plots were generated to understand how these genes may relate to specific biological processes (32). NP1 expressed genes associated with the gene ontology (GO) terms for endoplasmic reticulum and negative regulation of cell death, like HSPA5 (avg_logFC = 2.92), HSP90B1 (avg_logFC = 2.68), and SELENOS (avg_logFC = 2.09) (Fig. 4D). The expression of CRYAB (avg_logFC = 1.44), GADD45A (avg_logFC = 0.92), ATF4 (avg_logFC = 0.90), URI1 (avg_logFC = 0.85) and SFRP1 (avg_logFC = 0.83) in NP2 was associated with the GO term negative regulation of macromolecule biosynthetic processes (Fig. 4E). NP3 was defined by genes relating to the GO term for tissue development, like MUSTN1 (avg_logFC = 1.56), ECRG4 (avg_logFC = 1.39), SPP1 (avg_logFC = 1.20), CLEC3B (avg_logFC = 1.05), RHOC (avg_logFC = 0.97), IGFBP5 (avg_logFC = 0.97), S100A10 (avg_logFC = 0.96), CNMD (avg_logFC = 0.92), INHBA (avg_logFC = 0.90), MGP (avg_logFC = 0.88), MSN (avg_logFC = 0.81), HSPB1 (avg_logFC = 0.79), CAV1 (avg_logFC = 0.74), ACTG1 (avg_logFC = 0.73) and CLIC1 (avg_logFC = 0.72) (Fig. 4F). The top DEGs in NP4 were associated with the GO terms for the endomembrane system, positive regulation of cell communication and positive regulation of signal transduction. These genes included APOD (avg_logFC = 1.61), HMOX1 (avg_logFC = 0.95), CHI3L1 (avg_logFC = 0.95), S100A4 (avg_logFC = 0.87), GJA1 (avg_logFC = 0.85) and LAMTOR5 (avg_logFC = 0.80) (Fig. 4G). The last subpopulation of NP cells, NP5, was distinguished by an upregulation of canonical NP extracellular matrix (ECM) proteins, such as COL2A1 (avg_logFC = 2.24) and COMP (avg_logFC = 1.25). Expression of these genes was associated with the GO terms for endochondral bone morphogenesis and skeletal system morphogenesis GO terms (Fig. 4H). Based on these gene expression patterns, we hypothesize that clusters NP1, NP2 and NP4 are involved in NP cell survival, while clusters NP3 and NP5 are critical for NP tissue development and maintenance.
3.4. Analysis of the oAF Partition and oAF Cell Subpopulations
To uncover novel phenotypic biomarkers of oAF cells, we compared the pooled gene expression of all oAF clusters to all other clusters, analogously to our analysis of the NP partition. We found that FBLN1, SERPING1, COL1A1, LUM, IGFBP6, COL1A2, CTSK, LGALS1 and CCN3 were the top DEGs in this partition based avg_logFC and difference value (Fig. 5A). These genes were significantly expressed in the oAF clusters with limited expression in the iAF and NP partitions. Additionally, IGFBP6, CTSK, LGALS1 and CCN3 were novel DEGs for the oAF population.
Comparison of the gene expression profiles for clusters oAF1, oAF2 and oAF3 within the oAF partition identified distinct functional roles for each subpopulation (Fig. 5B) Top DEGs in each cluster were identified (Fig. 5C) and analyzed using GSEA and Cnet plots, as with the NP analysis. oAF1 expressed genes associated with the GO term for cellular response to stimulus, such as GADD45B (avg_logFC = 4.35), ZFP35 (avg_logFC = 4.04), JUN (avg_logFC = 3.84), JUNB (avg_logFC = 3.63), ID2 (avg_logFC = 3.60), FOS (avg_logFC = 3.43), ID3 (avg_logFC = 3.42), EGR1 (avg_logFC = 3.33), MT2A (avg_logFC = 3.09), GADD45A (avg_logFC = 2.94), UBB (avg_logFC = 2.61), RND1 (avg_logFC = 2.58) and MT1A (avg_logFC = 2.56) (Fig. 5D). oAF2 was associated with the extracellular matrix GO term due to expression of SPARC (avg_logFC = 2.91), MMP2 (avg_logFC = 1.98), ASPN (avg_logFC = 1.49), PRELP (avg_logFC = 1.56), MFAP4 (avg_logFC = 1.47), FMOD (avg_logFC = 1.47), BGN (avg_logFC = 1.43), APOE (avg_logFC = 1.07) and THBS4 (avg_logFC = 1.06). This cluster was also noted by upregulation of canonical oAF collagens, like COL1A1 (avg_logFC = 5.23) and COL1A2 (avg_logFC = 5.07) (Fig. 5E). The last subpopulation of oAF cells, oAF3, was defined by an upregulation of genes associated with the GO term for anatomical structure formation involved in morphogenesis. Specifically, these included SPRY2 (avg_logFC = 1.03), TXNRD1 (avg_logFC = 0.91), SARS1 (avg_logFC = 0.82), COL4A1 (avg_logFC = 0.77) and GLUL (avg_logFC = 0.76) (Fig. 5F). Based on these gene expression patterns, we hypothesize that oAF1 is involved in AF cell survival, while clusters oAF2 and oAF3 are critical for AF tissue development.
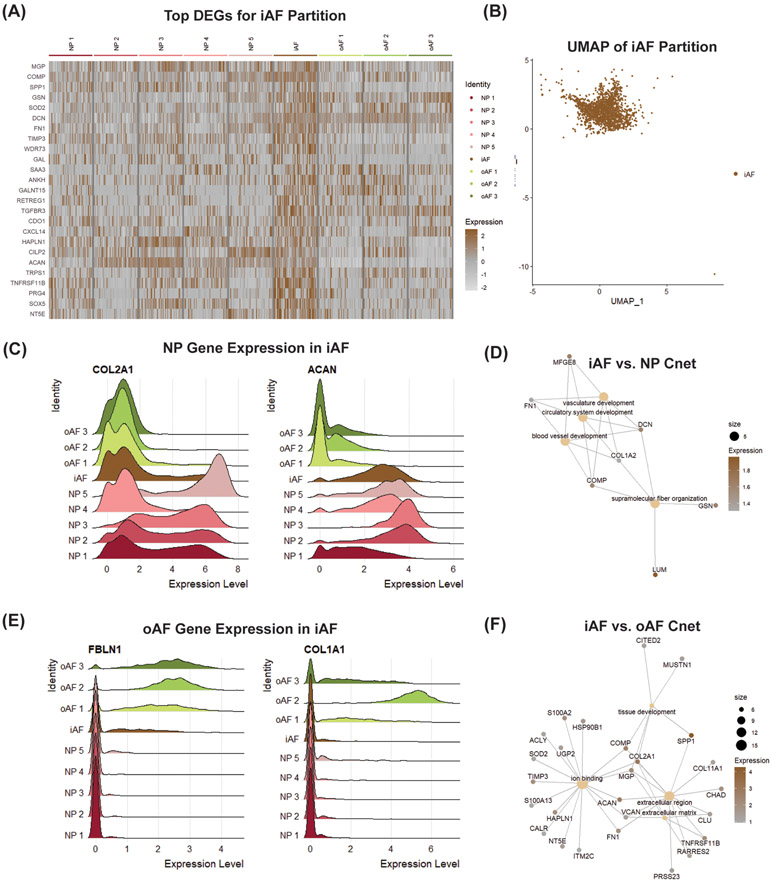
3.5. Distinguishing the iAF from oAF and NP Partitions
The iAF is an interesting tissue within the IVD because it originates from the sclerotome, like the oAF, but has a transcriptional profile that resembles the notochord-derived NP (35). To elucidate unique biomarkers for iAF cells, we compared the gene expression of the iAF partition to the NP and oAF partitions (Fig. 6A & B). Unbiased comparison of the gene signature for the iAF cluster compared to the pooled gene expression of the NP and oAF clusters showed that MGP, COMP, SPP1, GSN, SOD2, DCN, FN1, TIMP3, WDR73 and GAL were the top 10 DEGs in the iAF. Although these markers were fairly unique to the iAF cluster, a high percentage of cells in the iAF partition express the canonical NP markers COL2A1 (77.9%) and ACAN (94.6%) (Fig. 6C). Interestingly, the Cnet plot derived from top DEGs between the iAF and NP partitions showed that the iAF cluster upregulates GO terms associated with vasculature, circulatory and blood vessel development (Fig. 6D). These terms were driven by differential expression of iAF markers like DCN (avg_logFC = 1.61), COMP (avg_logFC = 1.58) and FN1 (avg_logFC = 1.34), but also by the expression oAF markers like LUM (avg_logFC = 1.96) and COL1A2 (avg_logFC = 1.31). iAF cells also expressed oAF markers FBN1 (74.0%) and COL1A1 (15.8%) (Fig. 6E). Analogously to the iAF vs. NP Cnet plots, the GO terms in the iAF vs. oAF Cnet plots were driven by differential expression of iAF markers like SPP1 (avg_logFC = 4.26), MGP (avg_logFC = 2.14), TIMP3 (avg_logFC = 1.97), FN1 (avg_logFC = 1.88) and SOD2 (avg_logFC = 1.04), and markers of NP subpopulations associated with tissue development, like COL2A1 (avg_logFC = 2.99), COMP (avg_logFC = 2.47) and MUSTN1 (avg_logFC = 1.36). Using Cnet plots to visualize linkages of DEGs to biological processes, the iAF was shown to upregulate genes associated with the GO terms for tissue development and the extracellular matrix (Fig. 6F). These results highlight the fibrocartilaginous nature of the iAF, it’s overlapping gene expression with the NP and oAF partitions, it’s potential role in ECM maintenance, and a novel role involving regulation of vascular system GO terms.
Figure 6: scRNA-seq analysis finds novel iAF cell marker genes.
(A) DEGA between iAF cluster and pooled NP and oAF partitions. (B) UMAP of iAF partition. Ridge plots showing overlapping gene expression of iAF cluster with (C) NP and (E) oAF partitions. Cnet plot displaying linkages of DEGs and associated biological processes between (D) iAF vs. NP and (F) iAF vs. oAF. Gene expression is the scaled expression value for the specified gene across individual cells.
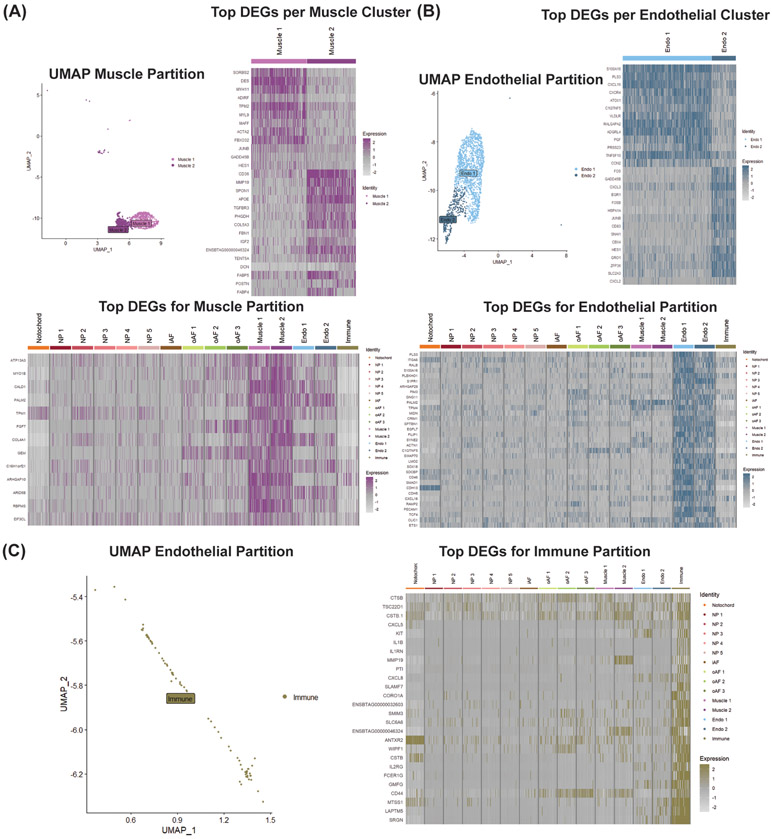
3.6. Analysis of Additional IVD Cell Populations
In addition to the predicted partitions (i.e., NP, oAF, iAF and notochord), our scRNA-seq analysis resolved partitions of muscle, endothelial and immune cells (Fig. 2B). Clusters in the muscle partition were defined by high expression of CALD1, TMP1 and RBPMS, which are common markers of smooth muscle cells (Fig. 2C & 7A). Muscle1 also upregulated ACTA2 and DES, marking this cluster as a population of myofibroblasts (Fig. 2C & 7A). Endothelial clusters were designated by the robust expression of canonical endothelial markers S1PR1, LMO2, CDH5 and PECAM1 (Fig. 2C and 7B). Upregulation of SRGN, SLAMF7, IL1B and KIT, which are all common markers of hematopoietic cells, in the immune cluster justified its annotation (Fig 2C & 7C). The presence of smooth muscle, endothelial and immune cell populations within our scRNA-seq analysis likely indicates that blood vessels were processed with IVDs for single-cell isolation, or that the IVD tissue system contains muscle, endothelial and immune cell populations.
Figure 7: scRNA-seq analysis discovers novel IVD cell types.
(A) UMAP of muscle partition, DGEA between muscle subpopulations compared to one another, and DGEA between pooled muscle subpopulations and all other clusters. (B) UMAP of endothelial partition, DGEA between endothelial subpopulations compared to one another, and DGEA between pooled endothelial subpopulations and all other clusters. (C) UMAP of immune partition and DGEA between Immune cluster and all other clusters. Expression value represents scaled expression of the specified gene on an individual cell barcode ID.
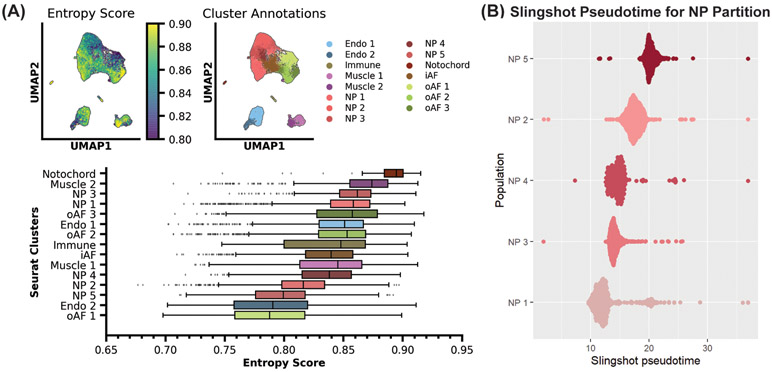
3.7. Presence of IVD Progenitor Cells
Understanding the function of tissue-specific progenitor cells is important for developing regenerative therapies; therefore, we were interested in searching for resident progenitor cells in our scRNA-seq dataset. We employed single-cell transcriptional entropy to estimate the stemness of cell clusters within our dataset. Using this analysis pipeline, we found that our notochord population had the highest transcriptional entropy score of 0.89, which correlates to being the most stem-like population (Fig. 8A) (53). The notochord is an embryonic structure that develops during gastrulation and notochordal cells are highly stem-like (54). Thus, the accurate identification of notochord cells as the most stem-like cluster in our population provided further confidence that entropy analysis could be used to identify potential progenitor cells within our dataset. Using this tool, we identified clusters NP3 and NP1 as having similarly high transcriptional entropy scores of 0.86. These were the highest scores for the NP partition, suggesting these clusters may contain resident NP progenitor cells. Clusters NP4, NP2 and NP5 had lower transcriptional entropy scores of 0.84, 0.82 and 0.80, respectively, suggesting these are more differentiated NP subpopulations. Within the oAF partition, we identified clusters oAF3 and oAF2 as having transcriptional entropy scores of 0.86 and 0.85 respectively. Cluster oAF1 had a transcriptional entropy score of 0.79, the lowest score of all clusters. This suggests that clusters oAF3 and oAF2 may contain resident oAF progenitor cells.
Figure 8: Entropy and pseudotime analyses identify stem-like populations of cells within IVD.
(A) Single-cell entropy analysis of all resolved clusters. Transcriptional entropy scores range from 0 to 1. High scores are correlated with stem-like cell populations and low scores are associated with differentiated cell populations. (B) Pseudotime analysis showing predicted differentiation trajectories of NP subpopulations. Clusters with lower pseudotime values are considered to be less differentiated than clustered with greater pseudotime values.
Slingshot pseudotime analysis was used as an additional tool to validate potential IVD progenitor cell populations. This tool created a pseudotime lineage score for individual cells to rank populations by differentiation timepoint. Through this analysis pipeline, it was found that subpopulation NP1 was the most stem-like cluster in the NP partition. It then ranked the NP clusters by average pseudotime score predicting NP1, NP3, NP4, NP2 and NP5 as the most progenitor-like to the most differentiated (Fig. 8B). These results, using an orthogonal analysis pipeline, corroborate our entropy analysis and further highlight clusters NP3 and NP1 as clusters with more progenitor-like populations. To generate pseudotime lineages, phylogenetic pseudotime algorithms rely on a continuum of data points to identify a trajectory (55). It is well established that the notochord differentiates into the NP (8); however, this differentiation trajectory could not be represented through this pseudotime analysis. We believe this is due to distance in differentiation time points between notochord and NP cells, with a lack of detection of the intermediary lineage populations.
4. Discussion
The poor understanding of heterogeneous cell populations within the IVD presents a barrier to deeper understanding of IVD pathophysiology and development of improved regenerative therapies. Previous studies, which employed bulk transcriptional profiling of NP and AF tissues (17-19), inherently masked IVD cellular heterogeneity by measuring average gene expression patterns. This study conducted scRNA-seq analysis of bovine caudal IVDs to elucidate the identity and function of heterogeneous cell populations within this tissue. Extensive bioinformatic analyses were employed to discover novel partitions, elucidate functional roles of subpopulations within partitions and identify potential progenitor cells. This deeper understanding of IVD cell types utilizing both unbiased and supervised analyses is needed to broadly advance IVD research, accelerate the development of novel therapies and reduce the global burden of discogenic back pain.
Cellular heterogeneity within the IVD has been of interest for many years, but there have only been three published IVD scRNA-seq papers that address this topic. Fernandes et al. used scRNA-seq to identify top DEGs between NP and AF tissue in human IVDs (28). Expression levels for these genes were measured in our bovine dataset and we found concordant gene expression in the NP partition for COL2A1, LPL, EFNB2 and RARRES2. We found limited expression of the top DEGs for the AF in our dataset. Comparing our results to Wang et al., who conducted scRNA-seq on rat caudal IVDs (29), we showed highly concordant expression for the classic NP (COL1A1, COL2A1 and ACAN), classic iAF (COL1A1, COL2A1, ACAN, SERPING, PRG4 and COL15A1) and classic oAF (COL1A1, COL2A1, ACAN, COL11A1 and SFRP2) markers they used to annotate their dataset. Interestingly, we also saw significant expression of some the novel iAF (MMP3 and INHBA) and novel oAF (MYOC, IGFBP5 and LUM) markers defined by Wang et al.. Looking more deeply at cellular heterogeneity, Wang and colleagues found similar levels of heterogeneity within the AF region of the IVD, but less heterogeneity within the NP region. Regardless, we identified concordant expression for the NP (UCP2 and ICAM1), iAF (MGP and MMP3), oAF (IGFBP5, CILP2, CLEC3B, SFRP4 and ASPN) subclusters. Calió et al. were the most recent group to explore IVD cell heterogeneity with scRNA-seq, using bovine caudal IVDs (30). Calió and colleagues found substantially greater heterogeneity in the NP and AF regions (27 and 24 clusters, respectively), which is likely due to the greater number of cells sequenced and resolution of clustering. However, our results aligned with those of Calió et al. in that we both identified clusters responsible for cellular response to stress (e.g., heat shock proteins) and ECM synthesis (e.g, collagens and small leucine rich proteoglycans). Overall, currently published IVD scRNA-seq papers corroborate our results, and differences can most likely be explained by species-level differences in gene expression and potential differences in methods for cell isolation and sequencing. For example, Fernandes et al. cultured their IVD cells prior to scRNA-seq, which could impact cell phenotype.
Cells in all clusters were annotated using a combination of supervised and unbiased annotation methods. Supervised cell population annotations were conducted using known marker gene expression and our cell type predictions were validated using a custom unbiased annotation tool. The annotation pipeline proved to be a powerful tool for assisting in the annotation of novel scRNA-seq datasets. We note that the pipeline forces cell identification toward the closest cell type within the training dataset, and is limited by the availability of published datasets. Since there are almost no publicly available scRNA-seq datasets for the IVD, cells in the NP partition were annotated as chondrocytes and cartilage cells, while cells in the oAF partition were annotated as fibroblasts. As the orthopaedics community continues to apply single-cell technologies to musculoskeletal tissues, additional publicly available datasets will be generated and future analysis tools, like our unbiased annotation pipeline, will be able to more accurately identify musculoskeletal cell types with greater specificity.
Novel phenotypic markers characterizing the NP and oAF cells were identified through analyses of the pooled gene signatures of the NP and oAF partitions. In the NP, CP, S100B, H2AC18, SNORC, CRELD2, PDIA4, DNAJC3, CHCHD7 and RCN2 were among the top DEGs, but have not been previously identified as NP marker genes. The CP gene encodes ceruloplasmin, a serum ferroxidase that utilizes copper to couple substrate oxidation with the four-electron reduction of oxygen to water (56). There is a dearth of information about H2AC18 expression; therefore, further research on this gene and the H2A histone-associated protein it encodes would be interesting for the IVD field. S100B (57), SNORC (58) and CRELD2 (59) are genes previously identified in articular chondrocytes, and associated with chondrocyte differentiation and phenotype maintenance. PDIA4 (60), DNAJC3 (61) and RCN2 (62) encode proteins localized to the endoplasmic reticulum (ER) and thought to be associated with ER stress. Upregulation of PDIA4 has been associated with IVD degeneration; therefore, further investigation of this gene may be interesting for the IVD field (63). CHCHD7 encodes a widely expressed protein with a poorly understood function, but genetic variation in the chromosome domain encompassing PLAG1-CHCHD7 influences bovine stature (64).
In the oAF, IGFBP6, CTSK, LGALS1 and CCN3 were identified as novel DEGs. IGFBP6 and CCN3, encode insulin-like growth factor (IGF) binding proteins that are important for modulating IGF signaling (65, 66). IGF signaling is important for IVD development (67) and polymorphisms of the IGFBP6 gene have been associated with IVD degeneration (68); supporting further investigations of these genes in IVDs. Previous studies of CTSK, which encodes the interstitial collagenolytic enzyme cathepsin K (69), found that the protein is localized to degenerated AF fibers (70), suggesting a role in IVD degeneration. LGALS1 encodes Galectin-1, a secreted lectin that can regulate proliferation, differentiation, migration and death of mesenchymal cells (71). In IVD degeneration, LGALS1 expression was found more in the NP than the AF (72), suggesting downregulation of this gene in the AF may be associated with degeneration. Discovery of these novel NP and oAF marker genes highlights the potential of this dataset and, more generally, scRNA-seq analyses to enhance our understanding of IVD biology.
scRNA-seq analysis resolved one iAF cluster that displayed significantly elevated expression of numerous genes compared to the pooled gene expression of the NP and oAF clusters. The top 10 DEGs for the iAF cluster were MGP, COMP, SPP1, GSN, SOD2, DCN, FN1, TIMP3, WDR73 and GAL. Expression of COMP and DCN have been previously associated with the iAF region of the IVD (73, 74), demonstrating the validity of our results. Interestingly, MGP and SPP1 were among the DEGs that distinguished cluster NP3 from the remaining NP clusters. This highlights the cartilaginous nature of the iAF and suggests that cells in NP3 may be transcriptionally similar to the fibrocartilaginous iAF. GSN encodes gelsolin, a very important actin-binding protein whose reduced expression has been correlated with patients suffering from rheumatoid arthritis (75). This may implicate a similar role of GSN in IVD degeneration. SOD2, FN1 and TIMP3 were all previously identified in the transformation of healthy NP cells to fibrocartilaginous NP cells during IVD degeneration (76-78). This shared gene expression between iAF and degenerated NP is supported by our network analysis comparisons of the iAF and NP partitions, which found that the iAF upregulated genes associated with the GO terms for vascular, circulatory and blood vessel development. Neovascularization is a hallmark of IVD degeneration (79); thus, the iAF may be a useful population of cells for studying IVD degeneration in the NP. To the authors’ knowledge, there are no published studies identifying the expression of WDR73 or GAL in the IVD, and these novel genes are worthy of further exploration. WDR73 encodes a protein involved in microtubule organization and dynamics during mitosis, and mutations in this gene have been associated with Galloway-Mowat syndrome (80). Galanin, encoded by GAL, is a neuropeptide involved in pain signaling (81). A previous study investigating neurochemical characteristics of IVD degeneration in a porcine model found that GAL was downregulated in sympathetic chain ganglia of pigs with IVD degeneration (82). Since markers that distinguish the iAF from the NP and oAF have been a longstanding knowledge gap in the field of IVD biology, the genes highlighted by our scRNA-seq analysis are an important step in understanding the role of the iAF in IVD homeostasis. Furthermore, iAF cells regulate genes important in neurovascular processes suggesting iAF may be important in modulation of discogenic pain processes.
Utilizing the highly resolved scRNA-seq dataset, we were able to identify 5 subpopulations of NP cells and 3 subpopulations of oAF cells, whose functional roles fell into two major categories: cell survival and tissue development. Based on the gene expression patterns of clusters NP1, NP2, NP4 and oAF1, we hypothesize that these are robust cell populations involved in cell survival under low oxygen tension and high mechanical loading. Cells in NP1 expressed SELENOS, SELENOK (83), HSP90B1 (84) and HSPA5 (85), which are all encode endoplasmic reticulum-associated proteins involved in protein folding, trafficking and degradation, and assist with cell survival decisions. GADD45A (86), CRYAB (87), URI1 (88), SFRP1 (89) and ATF4 (90) were all upregulated in NP2 cells, and these genes are related to cellular stress pathways. Upregulation of the PERK/ATF4/CHOP signaling pathway was shown to be related to IVD degeneration (91). The overexpression of APOD (92) and S100A4 (93), as in NP3, has been previously associated with oxidative stress and articular cartilage degeneration, respectively. NP4 cells also overexpress CHI3L and HMOX1, which are considered to be protective against oxidative stress-induced catabolism of the NP in the context of IVD degeneration (94, 95). Cells in oAF1 expressed a myriad of genes related to cellular stress pathways, including GADD45A, GADD45B, (86) UBB (96), FOS, JUN, JUNB (97), ID2, ID3 (98), MT2A and MT1A (99). Studies investigating the Fos/Jun signaling pathway in the IVD found opposing effects on IVD degeneration depending on which pathway activated it (100). Results therefore highlight a need for future studies to investigate these subpopulations of NP and oAF cells and better understand their molecular and cellular role in IVD homeostasis and degeneration.
GO terms associated with clusters NP3, NP5, oAF2 and oAF3 were related to tissue development, skeletal system morphogenesis, ECM and anatomical structure formation involved in morphogenesis, respectively, suggesting these populations are important for forming and maintaining the IVD structure. Cells in NP3 expressed MGP and SPP1, which regulate ectopic tissue calcification (101), MUSTN1, a gene expressed in musculoskeletal tissues during development (102), and CNMD, a protein that promotes chondrocyte differentiation and attenuates osteoarthritis progress by inhibiting catabolic activity (103, 104). COL2A1 and COMP, which encode ECM proteins canonically associated with the NP (9), were both upregulated in NP5. Cluster oAF2 was marked by the expression of canonical oAF ECM collagens, COL1A1 and COL1A2 (12), multiple small leucine-rich proteoglycans found in the oAF, like BGN, FMOD, ASPN (7, 105), and other proteins involved in collagen fibril assembly, like SPARC, MFAP4 and PRELP (106-108). Cells in oAF3 expressed COL4A1, a basement membrane-associated collagen (109). GLUL, SPRY2 and SARS1 were also upregulated in cluster oAF3, which are required for proper tissue development because they regulate cell proliferation and protein translation (110-112). For example, SPRY2 knockout mice show decreased skeletal size and trabecular bone mass, as a result of aberrant proliferation and impaired terminal differentiation (113). Cells phenotypically similar to subpopulations NP3, NP5, oAF2 and oAF3 may be promising for cell delivery because of upregulation of ECM-associated genes could result in ECM synthesis and structural regeneration of the IVD after implantation.
Using entropy and pseudotime analyses, we identified potential resident progenitor cells within the IVD, which may be involved in endogenous repair mechanisms and important for regenerative medicine research. This finding is highly significant because many studies address the presence of progenitor cells in the IVD; however, current findings mostly use in vitro approaches, with limited in vivo findings (11). High transcriptional entropy is associated with reduced focalized functions allowing for increased differentiation potential. Single-cell entropy analysis found that our notochord cluster was the most stem-like in our population, clusters NP3 and NP1 were the most stem-like subpopulations of the NP partition, and oAF3 and oAF2 were the most stem-like in the oAF partition. Slingshot pseudotime analysis corroborated entropy analysis results for clusters in the NP partition. Lineage tracing studies have demonstrated that the NP is derived from the notochord (114); thus, these analysis pipelines independently identified progenitor cells and their differentiation trajectories using orthogonal approaches. Furthermore, our analyses determined that notochordal cells can differentiate into multiple NP cell phenotypes. Future studies that isolate cells from clusters NP3 and NP1 are needed as a next step to confirm their stemness using in vitro and in vivo assays. Furthermore, future studies on oAF cell populations are needed to validate the stem-like characteristics of oAF3 and oAF2 suggested by the entropy analyses and further identify potential oAF progenitor populations.
Despite the numerous insights gathered from our scRNA-seq analysis of the IVD, it is not without limitations. First, our analysis used bovine caudal IVDs, due to the difficulty in obtaining fully intact, healthy human IVDs of sufficient quality. Bovine IVDs are commonly used as a large animal model for IVD research because of their similar mechanical loading, nutrient transport distances and gene expression to human IVDs (31). Additionally, we see concordant expression of NP and oAF genes identified through scRNA-seq of human IVD cells (28) in our bovine dataset. Therefore, we believe the marker genes and subpopulations of partitions analyzed in our dataset are likely to have implications for human IVDs, but additional species-level comparisons will be necessary to confirm these results. Second, IVD age is an important variable when studying resident cell populations, but we did not have precise control of the age of these specimens. The abattoir confirmed the age of our animals was between 2 to 3 years-old; thus, we are confident that age did not significantly bias our results. Future scRNA-seq experiments investigating how IVD cell populations change during development, maturation, aging and degeneration processes will require other model systems with more precise age control. Third, scRNA-seq analysis requires dissociation of the tissue of interest, resulting in a loss of spatial information about identified cell populations. Positional context is crucial for understanding how cells interact with one another during tissue development, homeostasis and degeneration; thus, corroboration of our dataset with spatial transcriptomics (115) would help resolve how IVD cell subpopulations interact with one another to maintain IVD function and is of interest for future research. Fourth, novel markers generated from our sequencing data were not validated with protein-level assays. We chose to focus this manuscript on the discovery and presentation of potential biomarkers since validated bovine antibodies are difficult to obtain and there were too many potential biomarkers discovered to be tested. Lastly, only sequencing data was used to identify potential progenitor cell populations and their differentiation trajectories. Future work will be required to assess the differentiation potential of identified progenitor cells and confirm the differentiation trajectories predicted by our analyses.
In conclusion, we identified numerous novel phenotypic biomarkers of the NP (CP, S100B, H2AC18, SNORC, CRELD2, PDIA4, DNAJC3, CHCHD7 and RCN2), oAF (IGFBP6, CTSK, LGALS1 and CCN3) and iAF (MGP, COMP, SPP1, GSN, SOD2, DCN, FN1, TIMP3, WDR73 and GAL) cells using scRNA-seq and inferred their relevance to the IVD function. Network analyses identified clusters involved in ECM maintenance and cell survival. Clusters NP3, NP5, oAF2 and oAF3, which were associated with ECM maintenance, are potentially desirable cells for exogenous cell therapies because they can promote tissue repair via ECM synthesis. Entropy and pseudotime analyses identified clusters notochordal, NP3, NP1, oAF3 and oAF2 as the most stem-like in their partitions, implying these clusters may contain interesting IVD-progenitor cells that could be targeted for endogenous repair strategies. Novel iAF marker genes were also identified to distinguish this partition from NP and oAF partitions, and transcripts suggest these cells could be important in degenerative processes. We believe these findings provide an exciting dataset and fundamental information that is broadly applicable to understand IVD cell physiology and function. Moreover, these findings will be crucial to inform novel cell repair strategies for treating IVD degeneration and reducing the global burden of discogenic back pain.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIH R01 AR057397 (JCI), NIH F31 AR077385 (CJP)]. Authors thanks Warren Hom, Associate Researcher, for assistance dissecting IVDs from bovine tails and Roosheel Patel, PhD Student, for important scientific discussions. Authors also thank Jill Gregory, Associate Director of the Levy Library Instructional Technology Group and Certified Medical Illustrator, for medical illustrations.
Nonstandard Abbreviations
- AF
Annulus fibrosus
- avg_logFC
log base 2-fold change
- DEGs
Differentially expressed genes
- DGEA
Differential gene expression analysis
- ECM
Extracellular matrix
- gseGO
Gene set enrichment analysis of gene ontology
- GO
Gene ontology
- iAF
Inner annulus fibrosus
- IVD
Intervertebral disc
- NP
Nucleus pulposus
- oAF
Outer annulus fibrosus
- PCA
Principal component analysis
- scRNA-seq
Single-cell RNA-sequencing
- UMAP
Unifold manifold approximation and projection
Footnotes
Conflict of Interest Statement
Authors have no conflicts of interest to disclose.
Data Sharing and Data Accessibility
The datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (GSE179714).
References
- 1.Hoy D, March L, Brooks P, Blyth F, Woolf A, Bain C, Williams G, Smith E, Vos T, Barendregt J, Murray C, Burstein R, and Buchbinder R (2014) The global burden of low back pain: estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis 73, 968–974 [DOI] [PubMed] [Google Scholar]
- 2.Dieleman JL, Cao J, Chapin A, Chen C, Li Z, Liu A, Horst C, Kaldjian A, Matyasz T, Scott KW, Bui AL, Campbell M, Duber HC, Dunn AC, Flaxman AD, Fitzmaurice C, Naghavi M, Sadat N, Shieh P, Squires E, and Murray CJL (2020) US Health Care Spending by Payer and Health Condition, 1996-2016. JAMA 323, 863–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J, Smeets RJ, Underwood M, and Lancet Low Back Pain Series Working Group. (2018) What low back pain is and why we need to pay attention. Lancet 391, 2356–2367 [DOI] [PubMed] [Google Scholar]
- 4.Ohtori S, Inoue G, Miyagi M, and Takahashi K (2015) Pathomechanisms of discogenic low back pain in humans and animal models. Spine J. 15, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 5.Smith LJ, Silverman L, Sakai D, Le Maitre CL, Mauck RL, Malhotra NR, Lotz JC, and Buckley CT (2018) Advancing cell therapies for intervertebral disc regeneration from the lab to the clinic: Recommendations of the ORS spine section. JOR Spine 1, e1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panebianco CJ, Meyers JH, Gansau J, Hom WW, and Iatridis JC (2020) Balancing biological and biomechanical performance in intervertebral disc repair: a systematic review of injectable cell delivery biomaterials. eCM 40, 239–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro IM and Risbud MV (2014) The intervertebral disc: Molecular and structural studies of the disc in health and disease. Springer, Wien [Google Scholar]
- 8.Risbud MV, Schaer TP, and Shapiro IM (2010) Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev. Dyn 239, 2141–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattappa G, Li Z, Peroglio M, Wismer N, Alini M, and Grad S (2012) Diversity of intervertebral disc cells: phenotype and function. J. Anat 221, 480–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes AJ, Isaacs MD, Hughes C, Caterson B, and Ralphs JR (2011) Collagen fibrillogenesis in the development of the annulus fibrosus of the intervertebral disc. Eur. Cell. Mater 22, 226–241 [DOI] [PubMed] [Google Scholar]
- 11.Séguin CA, Chan D, Dahia CL, and Gazit Z (2018) Latest advances in intervertebral disc development and progenitor cells. JOR Spine 1, e1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyre DR and Muir H (1976) Types I and II collagens in intervertebral disc. Interchanging radial distributions in annulus fibrosus. Biochem. J 157, 267–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bezci SE, Werbner B, Zhou M, Malollari KG, Dorlhiac G, Carraro C, Streets A, and O’Connell GD (2019) Radial variation in biochemical composition of the bovine caudal intervertebral disc. JOR Spine 2, e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoniou J, Steffen T, Nelson F, Winterbottom N, Hollander AP, Poole RA, Aebi M, and Alini M (1996) The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J. Clin. Invest 98, 996–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risbud MV, Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, Sakai D, and Hoyland JA (2015) Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J. Orthop. Res 33, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torre OM, Mroz V, Bartelstein MK, Huang AH, and Iatridis JC (2019) Annulus fibrosus cell phenotypes in homeostasis and injury: implications for regenerative strategies. Ann. N. Y. Acad. Sci 1442, 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CR, Sakai D, Nakai T, Toyama K, Mochida J, Alini M, and Grad S (2007) A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur. Spine J 16, 2174–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai D, Nakai T, Mochida J, Alini M, and Grad S (2009) Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine 34, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 19.Minogue BM, Richardson SM, Zeef LA, Freemont AJ, and Hoyland JA (2010) Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Akker GGH, Surtel DAM, Cremers A, Rodrigues-Pinto R, Richardson SM, Hoyland JA, van Rhijn LW, Welting TJM, and Voncken JW (2014) Novel immortal human cell lines reveal subpopulations in the nucleus pulposus. Arthritis Res. Ther 16, R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Akker GGH, Surtel DAM, Cremers A, Richardson SM, Hoyland JA, van Rhijn LW, Voncken JW, and Welting TJM (2016) Novel immortal cell lines support cellular heterogeneity in the human annulus fibrosus. PLoS ONE 11, e0144497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li K, Kapper D, Youngs B, Kocsis V, Mondal S, Kraus P, and Lufkin T (2019) Potential biomarkers of the mature intervertebral disc identified at the single cell level. J. Anat 234, 16–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Yan W, and Setton LA (2006) Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur. Spine J 15 Suppl 3, S303–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, and McCarroll SA (2015) Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, and Bielas JH (2017) Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rai MF, Wu C-L, Capellini TD, Guilak F, Dicks AR, Muthuirulan P, Grandi F, Bhutani N, and Westendorf JJ (2021) Single cell omics for musculoskeletal research. Curr. Osteoporos. Rep 19, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam V, Chen P, Yee A, Solis N, Klein T, Kudelko M, Sharma R, Chan WC, Overall CM, Haglund L, Sham PC, Cheah KSE, and Chan D (2020) DIPPER, a spatiotemporal proteomics atlas of human intervertebral discs for exploring ageing and degeneration dynamics. elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandes LM, Khan NM, Trochez CM, Duan M, Diaz-Hernandez ME, Presciutti SM, Gibson G, and Drissi H (2020) Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci. Rep 10, 15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Huang Y, Huang L, Shi K, Wang J, Zhu C, Li L, Zhang L, Feng G, Liu L, and Song Y (2020) Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthr. Cartil 29, 389–401 [DOI] [PubMed] [Google Scholar]
- 30.Calió M, Gantenbein B, Egli M, Poveda L, and Ille F (2021) The Cellular Composition of Bovine Coccygeal Intervertebral Discs: A Comprehensive Single-Cell RNAseq Analysis. Int. J. Mol. Sci 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demers CN, Antoniou J, and Mwale F (2004) Value and limitations of using the bovine tail as a model for the human lumbar spine. Spine 29, 2793–2799 [DOI] [PubMed] [Google Scholar]
- 32.Yu G, Wang L-G, Han Y, and He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019) Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018) Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Akker GGH, Koenders MI, van de Loo FAJ, van Lent PLEM, Blaney Davidson E, and van der Kraan PM (2017) Transcriptional profiling distinguishes inner and outer annulus fibrosus from nucleus pulposus in the bovine intervertebral disc. Eur. Spine J 26, 2053–2062 [DOI] [PubMed] [Google Scholar]
- 36.Rajpurohit R, Risbud MV, Ducheyne P, Vresilovic EJ, and Shapiro IM (2002) Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 308, 401–407 [DOI] [PubMed] [Google Scholar]
- 37.Melrose J, Smith SM, Fuller ES, Young AA, Roughley PJ, Dart A, and Little CB (2007) Biglycan and fibromodulin fragmentation correlates with temporal and spatial annular remodelling in experimentally injured ovine intervertebral discs. Eur. Spine J 16, 2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brun J, Lutz KA, Neumayer KMH, Klein G, Seeger T, Uynuk-Ool T, Wörgötter K, Schmid S, Kraushaar U, Guenther E, Rolauffs B, Aicher WK, and Hart ML (2015) Smooth Muscle-Like Cells Generated from Human Mesenchymal Stromal Cells Display Marker Gene Expression and Electrophysiological Competence Comparable to Bladder Smooth Muscle Cells. PLoS ONE 10, e0145153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herring BP, El-Mounayri O, Gallagher PJ, Yin F, and Zhou J (2006) Regulation of myosin light chain kinase and telokin expression in smooth muscle tissues. Am J Physiol, Cell Physiol 291, C817–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurdziel K, Vogt KR, Walton KD, Schneider GK, and Gumucio DL (2016) Transcriptome of the inner circular smooth muscle of the developing mouse intestine: Evidence for regulation of visceral smooth muscle genes by the hedgehog target gene, cJun. Dev. Dyn 245, 614–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayanagi T and Sobue K (2011) Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adh. Migr 5, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molino M, Woolkalis MJ, Prevost N, Praticó D, Barnathan ES, Taraboletti G, Haggarty BS, Hesselgesser J, Horuk R, Hoxie JA, and Brass LF (2000) CXCR4 on human endothelial cells can serve as both a mediator of biological responses and as a receptor for HIV-2. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1500, 227–240 [DOI] [PubMed] [Google Scholar]
- 43.Goncharov NV, Nadeev AD, Jenkins RO, and Avdonin PV (2017) Markers and biomarkers of endothelium: when something is rotten in the state. Oxid. Med. Cell. Longev 2017, 9759735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jha R, Singh M, Wu Q, Gentillon C, Preininger MK, and Xu C (2017) Downregulation of LGR5 Expression Inhibits Cardiomyocyte Differentiation and Potentiates Endothelial Differentiation from Human Pluripotent Stem Cells. Stem Cell Reports 9, 513–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andueza A, Kumar S, Kim J, Kang D-W, Mumme HL, Perez JI, Villa-Roel N, and Jo H (2020) Endothelial Reprogramming by Disturbed Flow Revealed by Single-Cell RNA and Chromatin Accessibility Study. Cell Rep. 33, 108491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahane N and Kalcheim C (2020) Neural tube development depends on notochord-derived sonic hedgehog released into the sclerotome. Development 147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulugulla SH, Packard TA, Galloway NLK, Grimmett ZW, Doitsh G, Adamik J, Galson DL, Greene WC, and Auron PE (2018) Distinct mechanisms regulate IL1B gene transcription in lymphoid CD4 T cells and monocytes. Cytokine 111, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortiz LA, Dutreil M, Fattman C, Pandey AC, Torres G, Go K, and Phinney DG (2007) Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proc Natl Acad Sci USA 104, 11002–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterling H, Saginario C, and Vignery A (1998) CD44 occupancy prevents macrophage multinucleation. J. Cell Biol 143, 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glowacka WK, Alberts P, Ouchida R, Wang J-Y, and Rotin D (2012) LAPTM5 protein is a positive regulator of proinflammatory signaling pathways in macrophages. J. Biol. Chem 287, 27691–27702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Zhang C, Zhang Z, Han B, Shen Z, Li L, Liu S, Zhao X, Ye F, and Zhang Y (2018) Specific clinical and immune features of CD68 in glioma via 1,024 samples. Cancer Manag. Res 10, 6409–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korpetinou A, Skandalis SS, Labropoulou VT, Smirlaki G, Noulas A, Karamanos NK, and Theocharis AD (2014) Serglycin: at the crossroad of inflammation and malignancy. Front. Oncol 3, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teschendorff AE and Enver T (2017) Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat. Commun 8, 15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corallo D, Trapani V, and Bonaldo P (2015) The notochord: structure and functions. Cell. Mol. Life Sci 72, 2989–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellman NE and Gitlin JD (2002) Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 22, 439–458 [DOI] [PubMed] [Google Scholar]
- 57.Diaz-Romero J and Nesic D (2017) S100A1 and S100B: Calcium Sensors at the Cross-Roads of Multiple Chondrogenic Pathways. J. Cell. Physiol 232, 1979–1987 [DOI] [PubMed] [Google Scholar]
- 58.Heinonen J, Taipaleenmäki H, Roering P, Takatalo M, Harkness L, Sandholm J, Uusitalo-Järvinen H, Kassem M, Kiviranta I, Laitala-Leinonen T, and Säämänen AM (2011) Snorc is a novel cartilage specific small membrane proteoglycan expressed in differentiating and articular chondrocytes. Osteoarthr. Cartil 19, 1026–1035 [DOI] [PubMed] [Google Scholar]
- 59.Wilson R, Golub SB, Rowley L, Angelucci C, Karpievitch YV, Bateman JF, and Fosang AJ (2016) Novel elements of the chondrocyte stress response identified using an in vitro model of mouse cartilage degradation. J. Proteome Res 15, 1033–1050 [DOI] [PubMed] [Google Scholar]
- 60.Wang Z, Zhang H, and Cheng Q (2020) PDIA4: The basic characteristics, functions and its potential connection with cancer. Biomed. Pharmacother 122, 109688. [DOI] [PubMed] [Google Scholar]
- 61.Petrova K, Oyadomari S, Hendershot LM, and Ron D (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludvigsen M, Jacobsen C, Maunsbach AB, and Honoré B (2009) Identification and characterization of novel ERC-55 interacting proteins: evidence for the existence of several ERC-55 splicing variants; including the cytosolic ERC-55-C. Proteomics 9, 5267–5287 [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Dai G, Wang L, Shang F, Jiang L, Li S, Huang L, Xia J, and Wei H (2019) Identification of key genes potentially related to intervertebral disk degeneration by microarray analysis. Genet. Test. Mol. Biomarkers 23, 610–617 [DOI] [PubMed] [Google Scholar]
- 64.Karim L, Takeda H, Lin L, Druet T, Arias JAC, Baurain D, Cambisano N, Davis SR, Farnir F, Grisart B, Harris BL, Keehan MD, Littlejohn MD, Spelman RJ, Georges M, and Coppieters W (2011) Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 65.Bach LA (2016) Current ideas on the biology of IGFBP-6: More than an IGF-II inhibitor? Growth Horm. IGF Res 30–31, 81–86 [DOI] [PubMed] [Google Scholar]
- 66.Katsube K, Sakamoto K, Tamamura Y, and Yamaguchi A (2009) Role of CCN, a vertebrate specific gene family, in development. Dev. Growth Differ 51, 55–67 [DOI] [PubMed] [Google Scholar]
- 67.Masuda K and An HS (2004) Growth factors and the intervertebral disc. Spine J. 4, 330S–340S [DOI] [PubMed] [Google Scholar]
- 68.Fu S, Lei W, Lanlan D, Qi-Ling Y, Liang L, Hanlin Z, Jianguo Z, and Yin-Gang Z (2017) Whole exome sequencing identified a novel IGFBP6 variant in a disc degeneration pedigree. Genet. Test. Mol. Biomarkers 21, 580–585 [DOI] [PubMed] [Google Scholar]
- 69.Salminen-Mankonen HJ, Morko J, and Vuorio E (2007) Role of cathepsin K in normal joints and in the development of arthritis. Curr. Drug Targets 8, 315–323 [DOI] [PubMed] [Google Scholar]
- 70.Ariga K, Yonenobu K, Nakase T, Kaneko M, Okuda S, Uchiyama Y, and Yoshikawa H (2001) Localization of cathepsins D, K, and L in degenerated human intervertebral discs. Spine 26, 2666–2672 [DOI] [PubMed] [Google Scholar]
- 71.He J and Baum LG (2006) Galectin interactions with extracellular matrix and effects on cellular function. Meth. Enzymol 417, 247–256 [DOI] [PubMed] [Google Scholar]
- 72.Jing L, So S, Lim SW, Richardson WJ, Fitch RD, Setton LA, and Chen J (2012) Differential expression of galectin-1 and its interactions with cells and laminins in the intervertebral disc. J. Orthop. Res 30, 1923–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ishii Y, Thomas AO, Guo XE, Hung CT, and Chen FH (2006) Localization and distribution of cartilage oligomeric matrix protein in the rat intervertebral disc. Spine 31, 1539–1546 [DOI] [PubMed] [Google Scholar]
- 74.Götz W, Barnert S, Bertagnoli R, Miosge N, Kresse H, and Herken R (1997) Immunohistochemical localization of the small proteoglycans decorin and biglycan in human intervertebral discs. Cell Tissue Res. 289, 185–190 [DOI] [PubMed] [Google Scholar]
- 75.Feldt J, Schicht M, Garreis F, Welss J, Schneider UW, and Paulsen F (2019) Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Rev. Mol. Med 20, e7. [DOI] [PubMed] [Google Scholar]
- 76.Zhou TY, Wu YG, Zhang YZ, Bao YW, and Zhao Y (2019) SIRT3 retards intervertebral disc degeneration by anti-oxidative stress by activating the SIRT3/FOXO3/SOD2 signaling pathway. Eur. Rev. Med. Pharmacol. Sci 23, 9180–9188 [DOI] [PubMed] [Google Scholar]
- 77.Yang F, Leung VYL, Luk KDK, Chan D, and Cheung KMC (2009) Injury-induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J. Pathol 218, 113–121 [DOI] [PubMed] [Google Scholar]
- 78.Tsuji T, Chiba K, Imabayashi H, Fujita Y, Hosogane N, Okada Y, and Toyama Y (2007) Age-related changes in expression of tissue inhibitor of metalloproteinases-3 associated with transition from the notochordal nucleus pulposus to the fibrocartilaginous nucleus pulposus in rabbit intervertebral disc. Spine 32, 849–856 [DOI] [PubMed] [Google Scholar]
- 79.Adams MA and Roughley PJ (2006) What is intervertebral disc degeneration, and what causes it? Spine 31, 2151–2161 [DOI] [PubMed] [Google Scholar]
- 80.Colin E, Huynh Cong E, Mollet G, Guichet A, Gribouval O, Arrondel C, Boyer O, Daniel L, Gubler M-C, Ekinci Z, Tsimaratos M, Chabrol B, Boddaert N, Verloes A, Chevrollier A, Gueguen N, Desquiret-Dumas V, Ferré M, Procaccio V, Richard L, and Antignac C (2014) Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway-Mowat syndrome. Am. J. Hum. Genet 95, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang P, Yu M, Shi M, Bo P, and Zhang Z (2020) Galanin peptide family regulation of glucose metabolism. Front. Neuroendocrinol 56, 100801. [DOI] [PubMed] [Google Scholar]
- 82.Coronel MF, Villar MJ, Brumovsky PR, and González SL (2017) Spinal neuropeptide expression and neuropathic behavior in the acute and chronic phases after spinal cord injury: Effects of progesterone administration. Peptides 88, 189–195 [DOI] [PubMed] [Google Scholar]
- 83.Yu S-S and Du J-L (2017) Selenoprotein S: a therapeutic target for diabetes and macroangiopathy? Cardiovasc. Diabetol 16, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ni M and Lee AS (2007) ER chaperones in mammalian development and human diseases. FEBS Lett. 581, 3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, Lee J, Liem D, and Ping P (2017) HSPA5 Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene 618, 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salvador JM, Brown-Clay JD, and Fornace AJ (2013) Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol 793, 1–19 [DOI] [PubMed] [Google Scholar]
- 87.Acunzo J, Katsogiannou M, and Rocchi P (2012) Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int. J. Biochem. Cell Biol 44, 1622–1631 [DOI] [PubMed] [Google Scholar]
- 88.Thomas PA, Mita P, Ha S, and Logan SK (2018) Role of the unconventional prefoldin proteins URI and UXT in transcription regulation. Adv. Exp. Med. Biol 1106, 85–94 [DOI] [PubMed] [Google Scholar]
- 89.Galli LM, Barnes T, Cheng T, Acosta L, Anglade A, Willert K, Nusse R, and Burrus LW (2006) Differential inhibition of Wnt-3a by Sfrp-1, Sfrp-2, and Sfrp-3. Dev. Dyn 235, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang H, Cai F, Zhang Y, Xue M, Liu L, Yang A, and Liu X (2017) Early-stage autophagy protects nucleus pulposus cells from glucose deprivation-induced degeneration via the p-eIF2α/ATF4 pathway. Biomed. Pharmacother 89, 529–535 [DOI] [PubMed] [Google Scholar]
- 91.Fujii T, Fujita N, Suzuki S, Tsuji T, Takaki T, Umezawa K, Watanabe K, Miyamoto T, Horiuchi K, Matsumoto M, and Nakamura M (2018) The unfolded protein response mediated by PERK is casually related to the pathogenesis of intervertebral disc degeneration. J. Orthop. Res 36, 1334–1345 [DOI] [PubMed] [Google Scholar]
- 92.Do Carmo S, Levros L-C, and Rassart E (2007) Modulation of apolipoprotein D expression and translocation under specific stress conditions. Biochim. Biophys. Acta 1773, 954–969 [DOI] [PubMed] [Google Scholar]
- 93.Yammani RR (2012) S100 proteins in cartilage: role in arthritis. Biochim. Biophys. Acta 1822, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang R, Xu C, Zhong H, Hu B, Wei L, Liu N, Zhang Y, Shi Q, Wang C, Qi M, Gu Y, Shen X, Tian Y, Liu Y, Cao P, Chen H, and Yuan W (2020) Inflammatory-sensitive CHI3L1 protects nucleus pulposus via AKT3 signaling during intervertebral disc degeneration. FASEB J. 34, 3554–3569 [DOI] [PubMed] [Google Scholar]
- 95.Yi W, Lan H, Wen Y, Wang Y, He D, Bai Z, Zhang Y, Jiang W, Liu B, Shen J, and Hu Z (2020) HO-1 overexpression alleviates senescence by inducing autophagy via the mitochondrial route in human nucleus pulposus cells. J. Cell. Physiol 235, 8402–8415 [DOI] [PubMed] [Google Scholar]
- 96.Welchman RL, Gordon C, and Mayer RJ (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 97.Karin M, Liu Z. g, and Zandi E (1997) AP-1 function and regulation. Curr. Opin. Cell Biol 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 98.Ling F, Kang B, and Sun X-H (2014) Id proteins: small molecules, mighty regulators. Curr. Top. Dev. Biol 110, 189–216 [DOI] [PubMed] [Google Scholar]
- 99.Ling X-B, Wei H-W, Wang J, Kong Y-Q, Wu Y-Y, Guo J-L, Li T-F, and Li J-K (2016) Mammalian Metallothionein-2A and Oxidative Stress. Int. J. Mol. Sci 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yokoyama K, Hiyama A, Arai F, Nukaga T, Sakai D, and Mochida J (2013) C-Fos regulation by the MAPK and PKC pathways in intervertebral disc cells. PLoS ONE 8, e73210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gopalakrishnan R, Suttamanatwong S, Carlson AE, and Franceschi RT (2005) Role of matrix Gla protein in parathyroid hormone inhibition of osteoblast mineralization. Cells Tissues Organs (Print) 181, 166–175 [DOI] [PubMed] [Google Scholar]
- 102.Hadjiargyrou M (2018) Mustn1: A Developmentally Regulated Pan-Musculoskeletal Cell Marker and Regulatory Gene. Int. J. Mol. Sci 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu S, Qiu H, Bennett S, Kuek V, Rosen V, Xu H, and Xu J (2019) Chondromodulin-1 in health, osteoarthritis, cancer, and heart disease. Cell. Mol. Life Sci 76, 4493–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takao T, Iwaki T, Kondo J, and Hiraki Y (2000) Immunohistochemistry of chondromodulin-I in the human intervertebral discs with special reference to the degenerative changes. Histochem. J 32, 545–550 [DOI] [PubMed] [Google Scholar]
- 105.Singh K, Masuda K, Thonar EJ-MA, An HS, and Cs-Szabo G (2009) Age-related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine 34, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bedore J, Leask A, and Séguin CA (2014) Targeting the extracellular matrix: matricellular proteins regulate cell-extracellular matrix communication within distinct niches of the intervertebral disc. Matrix Biol. 37, 124–130 [DOI] [PubMed] [Google Scholar]
- 107.Zhu S, Ye L, Bennett S, Xu H, He D, and Xu J (2021) Molecular structure and function of microfibrillar-associated proteins in skeletal and metabolic disorders and cancers. J. Cell. Physiol 236, 41–48 [DOI] [PubMed] [Google Scholar]
- 108.Li H, Yang HH, Sun ZG, Tang HB, and Min JK (2019) Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients. Bone Joint Res. 8, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kühn K (1995) Basement membrane (type IV) collagen. Matrix Biol. 14, 439–445 [DOI] [PubMed] [Google Scholar]
- 110.Bott AJ, Maimouni S, and Zong W-X (2019) The pleiotropic effects of glutamine metabolism in cancer. Cancers (Basel) 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guy GR, Jackson RA, Yusoff P, and Chow SY (2009) Sprouty proteins: modified modulators, matchmakers or missing links? J. Endocrinol 203, 191–202 [DOI] [PubMed] [Google Scholar]
- 112.Turanov AA, Xu X-M, Carlson BA, Yoo M-H, Gladyshev VN, and Hatfield DL (2011) Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv. Nutr 2, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joo A, Long R, Cheng Z, Alexander C, Chang W, and Klein OD (2016) Sprouty2 regulates endochondral bone formation by modulation of RTK and BMP signaling. Bone 88, 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCann MR, Tamplin OJ, Rossant J, and Séguin CA (2012) Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis. Model. Mech 5, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, and Frisén J (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.