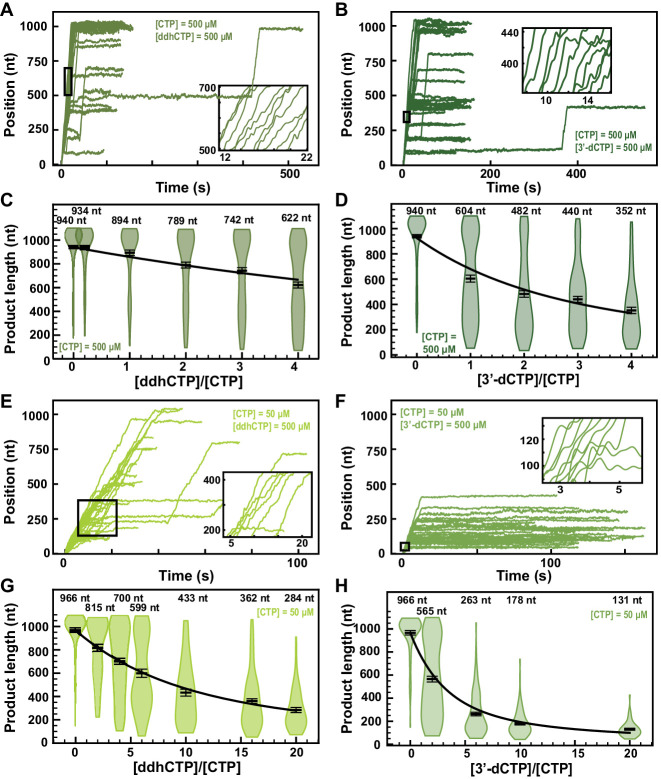
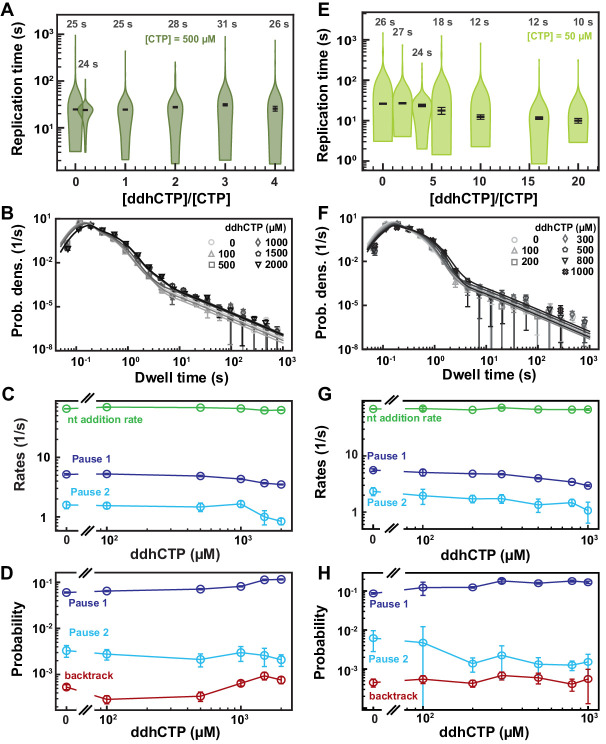
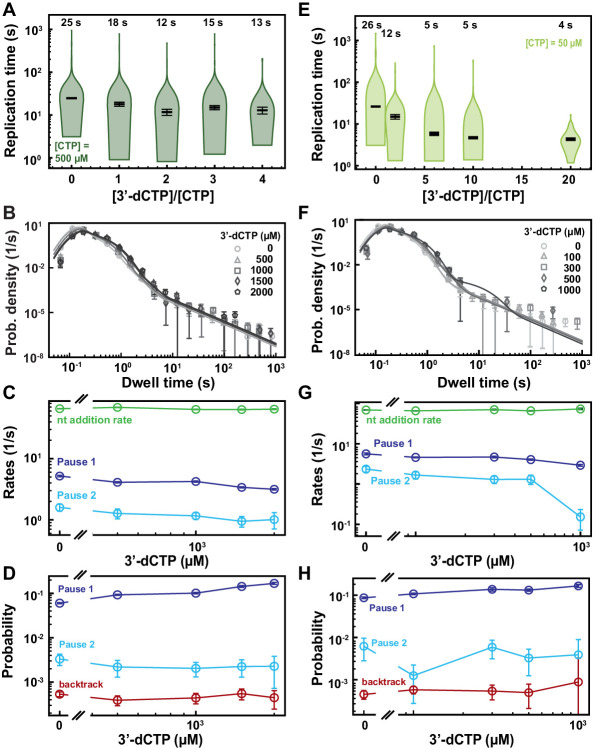
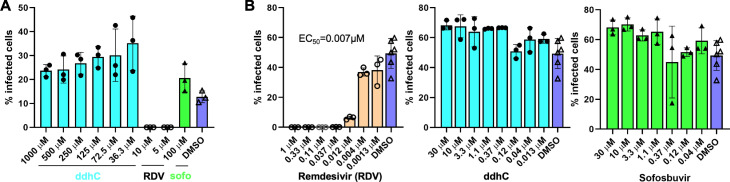
Figure 6. ddhCTP and 3ʹ-dCTP inhibit efficiently the SARS-CoV-2 polymerase.
(A, B) SARS-CoV-2 polymerase activity traces for 500 µM NTPs and 500 µM of either (A) ddhCTP or (B) 3ʹ-dCTP. (C, D) SARS-CoV-2 polymerase product length using the indicated concentration of CTP, 500 µM of other NTPs, as a function of either (C) [ddhCTP]/[CTP] or (D) [3ʹ-dCTP]/[CTP]. The mean values are indicated above the violin plots, and represented by horizontal black thick lines flanked by one standard deviation error bars extracted from 1000 bootstraps. (E, F) SARS-CoV-2 polymerase activity traces in the presence of 50 µM of CTP, 500 µM of all other NTPs, and 500 µM of either (E) ddhCTP or (F) 3ʹ-dCTP. (G, H) SARS-CoV-2 polymerase activity traces product length using 50 µM CTP, 500 µM of other NTPs, as a function of the stoichiometry of either (G) [ddhCTP]/[CTP] or (H) [3ʹ-dCTP]/[CTP]. The mean values are indicated above the violin plots, and represented by horizontal black thick lines flanked by one standard deviation error bars extracted from 1000 bootstraps. In (C, D, G, H), the solid lines are the fits of the terminator effective incorporation rate (see Materials and methods). In (A, B, E, F), the insets are a zoom-in of the replication traces captured in the black square.