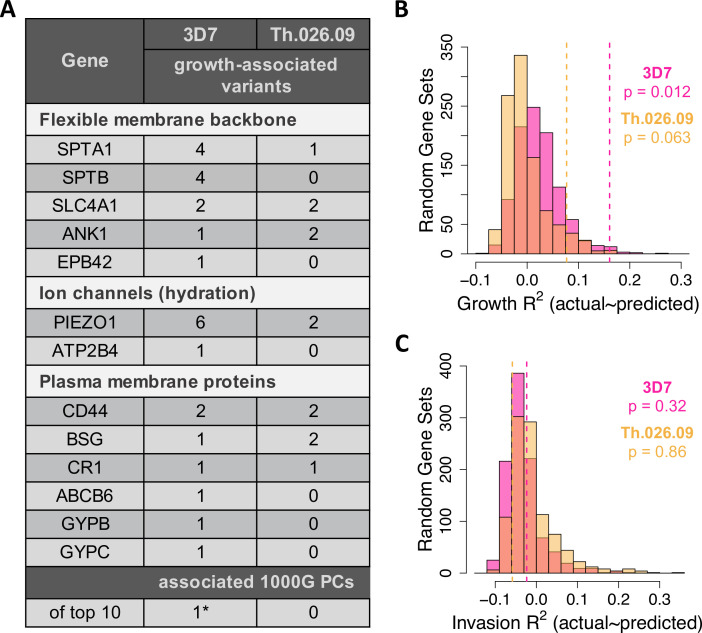
Figure 5. Common variation in malaria-associated genes predicts Plasmodium falciparum fitness in non-carrier RBCs.
(A) Variants in 23 malaria-related genes (Figure 5—source data 1) and genetic PCs selected by LASSO in at least >40% of train data sets. Each model was trained on ~90% of the measured data (B C) and tested on the remaining 10% (B C). The following genes had no associated variants in non-carriers: CD55, EPB41, FPN, G6PD, GYPA, GYPE, HBA1/2, HBB, and HP. *The only significant PC association was driven by a single East Asian donor (Figure 5—figure supplement 5). (B, C) Variance in parasite fitness explained by LASSO models including 23 malaria-related genes, the top 10 PCs, and RBC phenotypes. Dashed lines indicate average R2 for models using the measured test data. Each histogram shows R2 for models including variants from 23 random genes in the RBC proteome (Figure 5—source data 2) instead of malaria-related genes. All predictors with non-zero LASSO support are shown in Figure 5—source data 3. Additional histograms from permuted data are shown in Figure 5—figure supplement 1. The variance explained by variants undiscovered by previous GWAS is shown in Figure 5—figure supplement 4. GWAS, genome-wide association studies; PC, principal component; RBC, red blood cell.