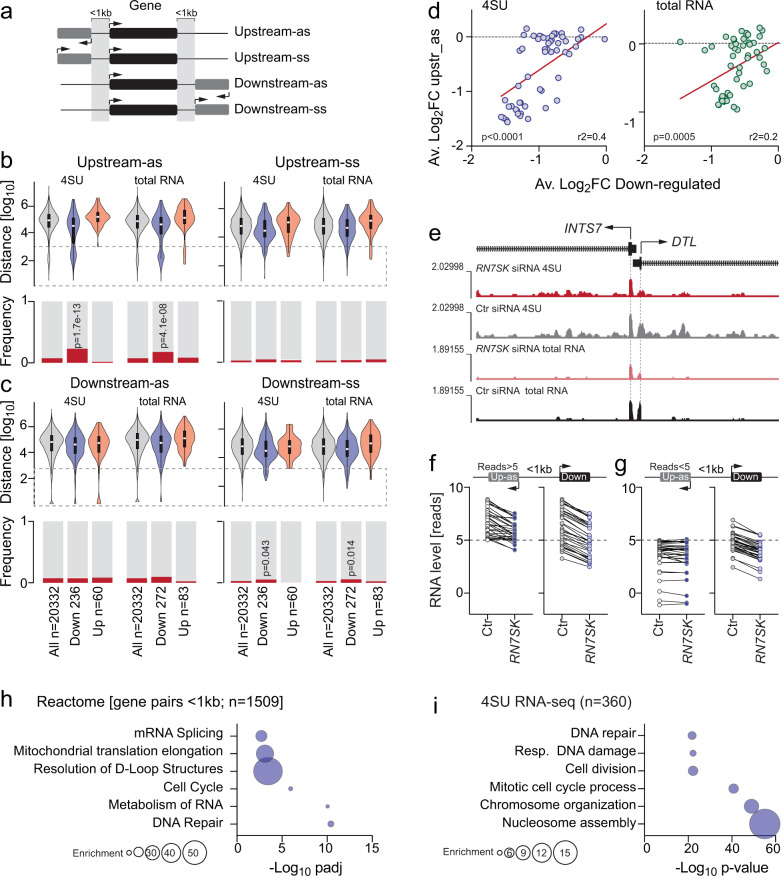
Fig. 4. 7SK orchestrates bi-directional transcription of highly expressed gene pairs.
a Illustration of the analyses shown in (b, c). b, c Distance (upper panels) and frequency (lower panels) of genes with upstream (b) or downstream (c) antisense (as) (left panels) or sense (ss) (right panels) genes within 1000 bases (1 kb) distance (grey dotted box). Up- (orange) and down- (blue) regulated genes were defined based on two independent 4SU and total RNA-seq datasets. All (grey) = all protein-coding genes; Up = genes with log2FC > 0.3 and padj < 0.05; Down = genes with log2FC < −0.3 and padj < 0.05 (Wald test with FDR correction). The Up and Down groups were compared to all protein-coding genes. Violin plots show the median and interquartile range and width indicating frequency (b, c upper panels). P-value calculation (see “Methods”). d Correlation of RNA levels of downregulated genes with their upstream as genes shown as average log2 fold-changes (FC) in two independent 4SU (left panel) and total RNA (right panel) sequencing datasets. P-value tests slope deviation from 0. e–g UCSC genome browser shots of bi-directional repressed DTL and its upstream anti-sense gene partner INTS7 (e). RNA levels (reads) of new transcripts of downregulated genes (padj < 0.05; FC < −0.5; n = 485) with bi-directional as gene less than 1 kb away and more than an average of 5 reads (n = 36 genes) (f) or less than 5 reads (n = 33 genes) (g) in upstream antisense (dark blue) or downstream sense (light blue) directions in control (ctr) siRNA transduced cells. h Reactome of all bi-directionally orientated gene pairs less than 1 kb apart. i Gene ontology analyses using all common significantly (padj < 0.5; Wald test with FDR correction) downregulated genes in the 4SU RNA seq datasets. Background: all expressed genes. Source data are provided as a Source Data file.