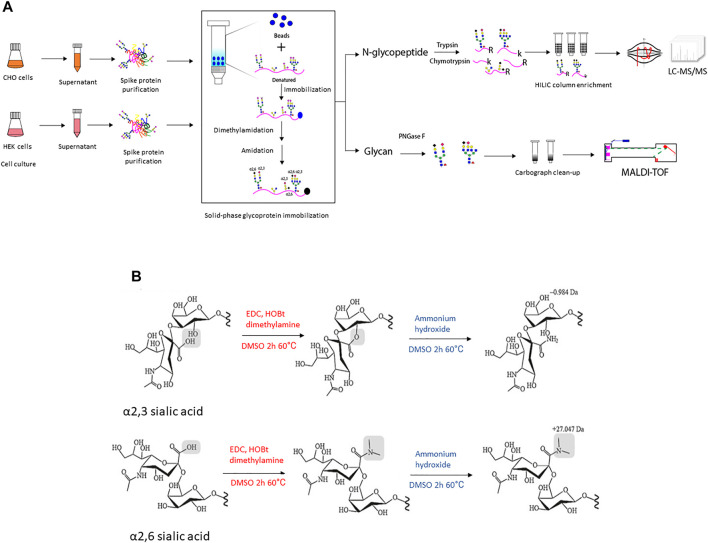
FIGURE 1.
(A) The illustration of experimental workflow in this project. Recombinant SARS-CoV-2 spike protein was transiently expressed in CHO and HEK cells in serum-free media, respectively. The conditioned supernatant was harvested and spike protein with a 6xhis tag at C-terminal was purified through Ni-NTA column and the protein purity was verified using SDS-PAGE followed by Coomassie blue staining (Supplementary Figure S2). Then equal amounts of purified recombinant spike protein were denatured and conjugated to an aldehyde-activated bead-based solid phase for N-glycan/N-glycopeptide analysis. The conjugated glycopeptides were subjected to reductive amination, lactonization and dimethylamidation for individual α2,3- and α2,6-linked sialic acids labeling. Subsequently, for N-glycopeptide analysis, trypsin and chymotrypsin digestion was performed and the flow-through digested N-glycopeptides were enriched using the HILIC column. The enriched N-glycopeptides were analyzed using LC-MS/MS mass spectrometry. Meanwhile, for N-glycan analyses of recombinant spike proteins from each cell line, PNGase F digested was applied to release N-glycans, followed with Carbograph column for N-glycan clean-up. Then N-glycans were analyzed using MALDI-TOF mass spectrometry. (B) The schematic illustration of two-step reactions in this project. The first step–dimethylamine treatment under EDC + HOBt condition, α2,3-sialic acid forms a lactone to the neighbor galactose, while α2,6-sialic acid forms a stable dimethyl amid structure. The second step-ammonium hydroxide treatment, α2,3-sialic acid further formed a stable amidation structure, while α2,6-sialic acid formed dimethyl amide remains stable in ammonium hydroxide treatment.