Fig. 6.
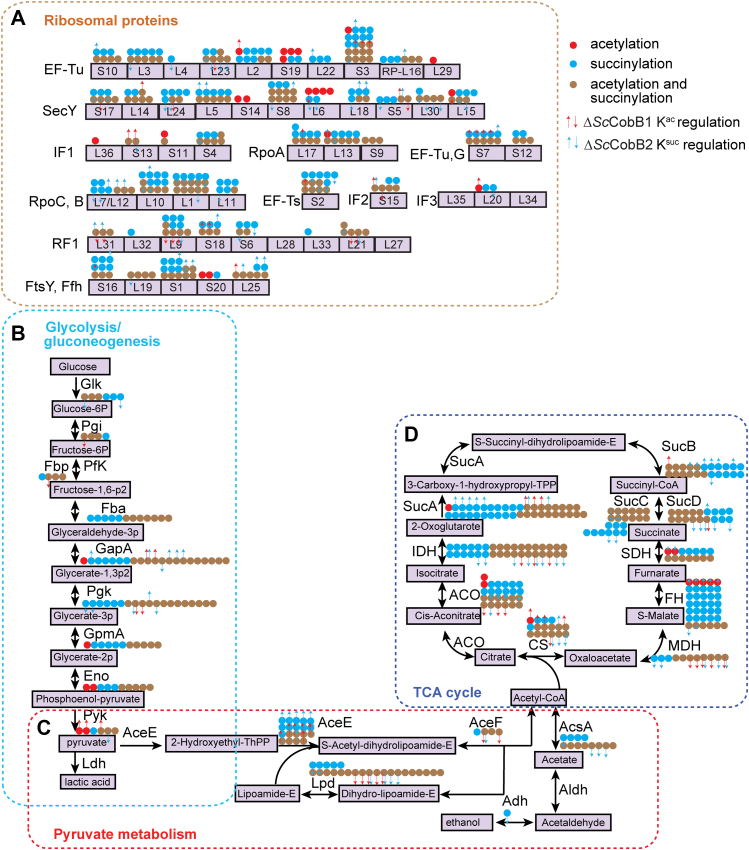
The enriched bimodified proteins are involved in protein translation and carbon metabolism pathways. The bimodified (acetylated and succinylated) proteins enriched in ribosomes (A), glycolysis/gluconeogenesis (B), pyruvate metabolism (C), and tricarboxylic acid (TCA) cycle (D) were mapped onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The acetylation-only sites are shown as red dots, succinylation-only sites are shown as blue dots, and bimodified sites are shown as brown dots. The modified sites in response to knocking out of SccobB1 or SccobB2 were quantified and mapped onto the KEGG pathways. The acetylation sites regulated by ΔSccobB1 and the succinylation sites regulated by ΔSccobB2 are shown in red and blue arrows, respectively. ACO, aconitate hydratase; AceE, pyruvate dehydrogenase E1 component; AceF, pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase); AcsA, acetyl-CoA synthetase; Adh, alcohol dehydrogenase; Aldh, aldehyde dehydrogenase; CS, citrate synthase; Eno, phosphopyruvate hydratase; Fba, fructose-bisphosphate aldolase; FH, fumarate hydratase; GapA, glyceraldehyde-3-phosphate dehydrogenase; Glk, glucokinase; GpmA, phosphoglycerate mutase; IDH, isocitrate dehydrogenase; Lpd, dihydrolipoamide dehydrogenase; MDH, malate dehydrogenase; N-resize, upregulated; PfkA, 6-phosphofructokinase; Pgi, glucose-6-phosphate isomerase; Pgk, phosphoglycerate kinase; Pyk, pyruvate kinase; S-resize, downregulated; SDH, succinate dehydrogenase; SucA, 2-oxoglutarate dehydrogenase E1 component; SucB, 2-oxoglutarate dehydrogenase E2 component; SucC, acetyl-CoA synthetase subunit beta; SucD, acetyl-CoA synthetase subunit alpha.