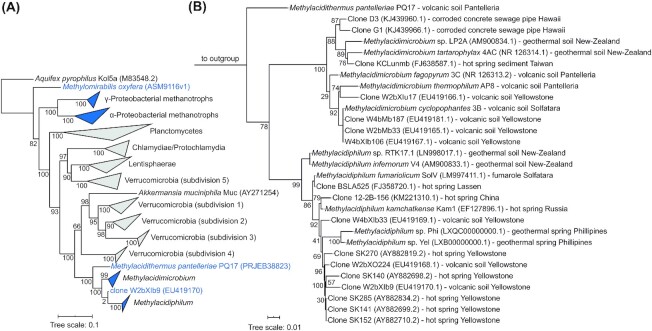
Figure 1.
(A) Overview 16S rRNA gene phylogenetic tree of Verrucomicrobia, Lentisphaerae, Chlamydiae, Planctomycetes and methanotrophic Alpha- and Gammaproteobacteria. Blue shading indicates species that are methanotrophic. Accession numbers are indicated between brackets. 16S rRNA gene alignment was constructed using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree that was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GTR-gamma. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. (B) Detailed 16S rRNA gene tree of the verrucomicrobial methanotrophs. 16S rRNA gene alignment of verrucomicrobial methanotrophs together with an outgroup of Planctomycetes constructed using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree that was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GTR-gamma. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. Isolation location and accession number are indicated after each strain.