Figure 2.
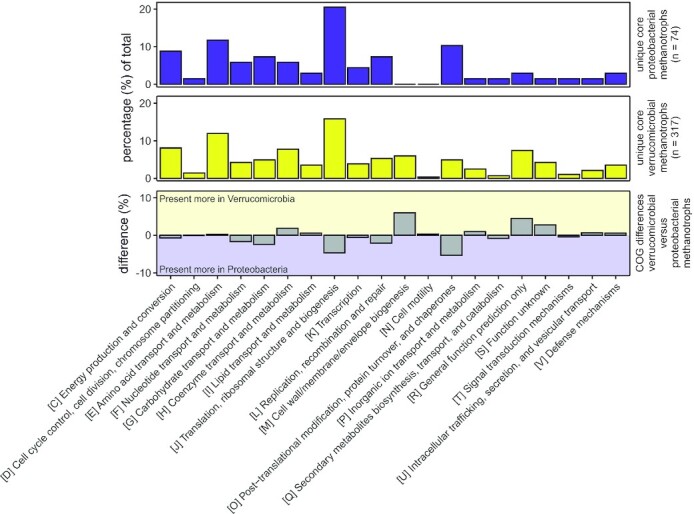
Bar graphs comparing the composition of Clusters of Orthologous Groups of proteins (COGS) of the unique core genomes of the verrucomicrobial methanotrophs and the proteobacterial methanotrophs. Core genomes were calculated by using usearch (Edgar 2010) with a cut-off value of 0.5 to group all genes into gene clusters. Clusters present in all genomes of the verrucomicrobial methanotrophs, as well as absent in all genomes of the proteobacterial methanotrophs, are defined as the unique core genome of verrucomicrobial methanotrophs. Clusters present in all genomes of the proteobacterial methanotrophs, as well as absent in all genomes of the verrucomicrobial methanotrophs, are defined as the unique core genome of proteobacterial methanotrophs. Analysis performed with 22 genomes in total, of which 11 verrucomicrobial methanotrophs: Methylacidiphilum fumariolicum SolV, Methylacidiphilum infernorum V4, Methylacidiphilum kamchatkense Kam1, Methylacidiphilum sp. Phi, Methylacidiphilum sp. Yel, Methylacidimicrobium cyclopophantes 3B, Methylacidimicrobium fagopyrum 3C, Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium thermophilum AP8, Methylacidimicrobium sp. LP2A and Methylacidithermus pantelleriae PQ17, and 11 proteobacterial methanotrophs: Methylobacterium extorquens AM1, Methylocaldum szegediense O-12, Methylocapsa acidiphila B2, Methylocella silvestris BL2, Methylococcus capsulatus Bath, Methylocystis rosea SV97T, Methyloferula stellata AR4, Methylomarinum vadi IT4, Methylomicrobium alcaliphilum 20Z, Methylomonas denitrificans FJG1 and Methylosinus trichosporium OB3b.
