Figure 3.
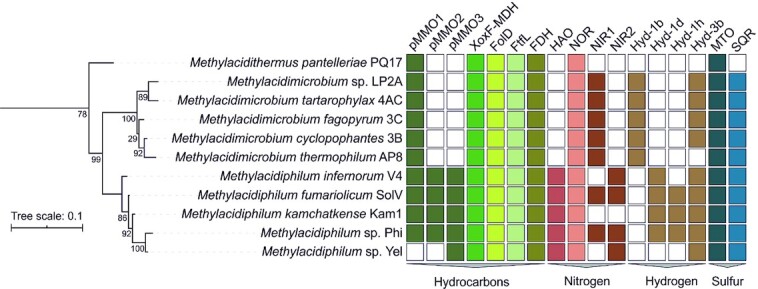
16S rRNA gene phylogenetic tree of verrucomicrobial methanotrophs and the presence or absence of genes involved in the oxidation of hydrocarbons, ammonia, hydrogen and sulfur compounds. Planctomycetes were used as an outgroup. Sequences were aligned using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GRT-GAMMA. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. Presence of genes of interest was examined using BLASTp and of each hit the amino acid sequence was manually inspected for domains and identity. The result table was transformed to a binary ITOL dataset and both the 16S tree and the table were visualised using ITOL v5.5.1. pMMO: particulate methane monooxygenase; XoxF-MDH: lanthanide-dependent methanol dehydrogenase; FolD: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase; FtfL: formate-tetrahydrofolate ligase; FDH: formate dehydrogenase; HAO: hydroxylamine oxidoreductase; NOR: nitric oxide reductase; NIR: nitrite reductase; Hyd: type of [NiFe] hydrogenase (based on Søndergaard, Pedersen and Greening 2016); MTO: methanethiol oxidase; SQR: sulfide:quinone oxidoreductase.
