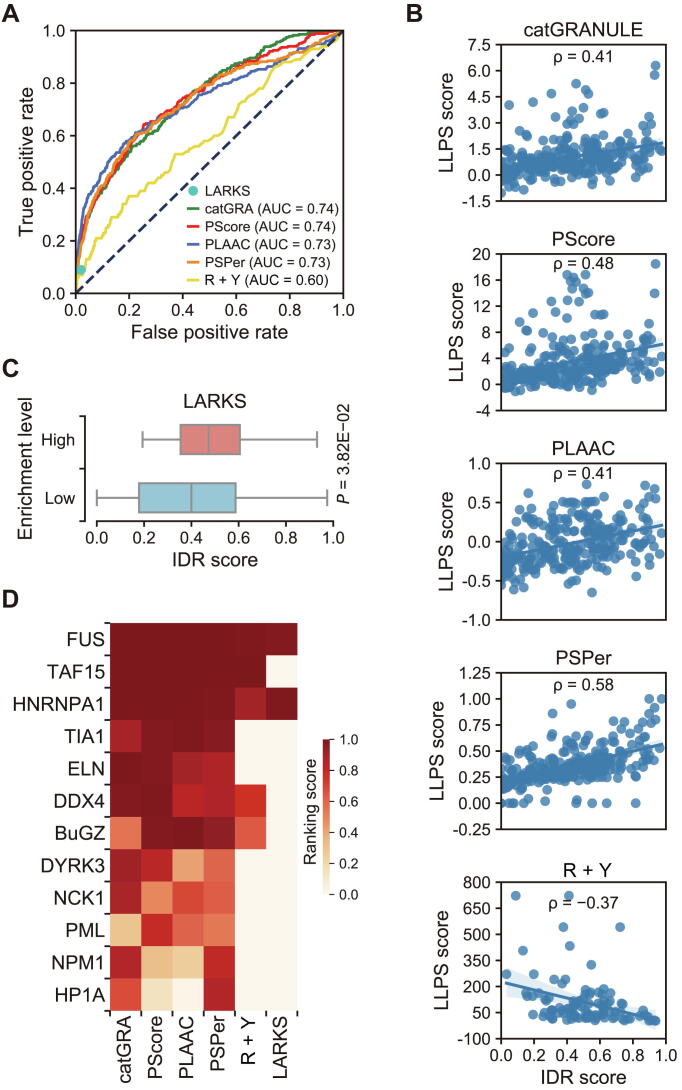
Figure 2.
Comparison of phase separation predictors on human proteome
A. ROC curve for each predictor. Since PScore, PLAAC, and PSPer have restrictions on the length of protein sequence, only catGRANULE returned scores for all human proteins. Except for catGRANULE, the AUC scores of the remaining tools were calculated on the subsets of P and N sets. Prediction of LARKS was shown as a point since it did not provide scores for each protein. B. Scatter plots of predicted values for proteins in set P, with one axis being the IDR score and the other axis being the phase separation score. Spearman correlation coefficient with P < 0.05 indicates significant correlation. C. Proteins in set P were divided into LARKS High and LARKS Low groups, according to whether the protein is in the list of LARKS-enriched proteins. P value is calculated through the two-sided Mann–Whitney U test. D. Ranking scores of 12 specific LLPS proteins by six phase separation predictors. ROC, receiver operating characteristic; AUC, area under the curve; LARKS, low-complexity aromatic-rich kinked segment; LLPS, liquid–liquid phase separation.