Figure 1.
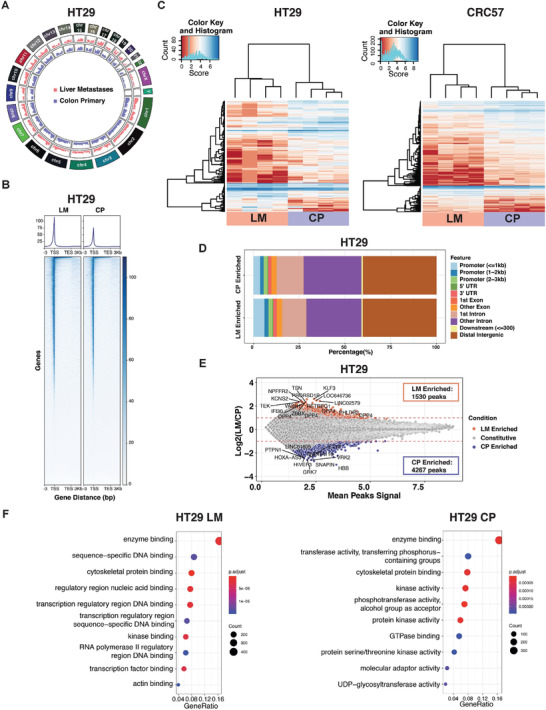
Epigenetic profiling on CRC liver metastases and CRC primary tumor. A) Circos plot revealing global chromatin accessibility change across different chromosomes in HT29 liver metastases versus primary CRC tumor. B) Heatmap for chromatin accessibility across gene body from TSS to transcription end site (TES) with 3 kb flanking regions in HT29 liver metastases versus primary CRC tumor. C) Hierarchical heatmap of 1000 chromatin accessible regions with the highest variance between liver metastases and primary CRC tumor in HT29 and CRC57 across four biological replicates. D) Distribution of enriched chromatin accessible regions across the genome in HT29 liver metastases versus primary CRC tumor. E) MA‐plot showing the differentially enriched (DE) accessible chromatin regions in HT29 liver metastases versus primary CRC tumor. F) Gene Ontology (GO) analysis on molecular functions for DE accessible chromatin regions in HT29 liver metastases versus primary CRC tumor. LM, liver metastases. CP, CRC primary tumor.
