Figure 1. In S. pombe, a deletion of the abo1 gene causes a marked cell proliferation defect suppressed by mutations in HIRA or CAF1 complexes or in histones H3 or H4.
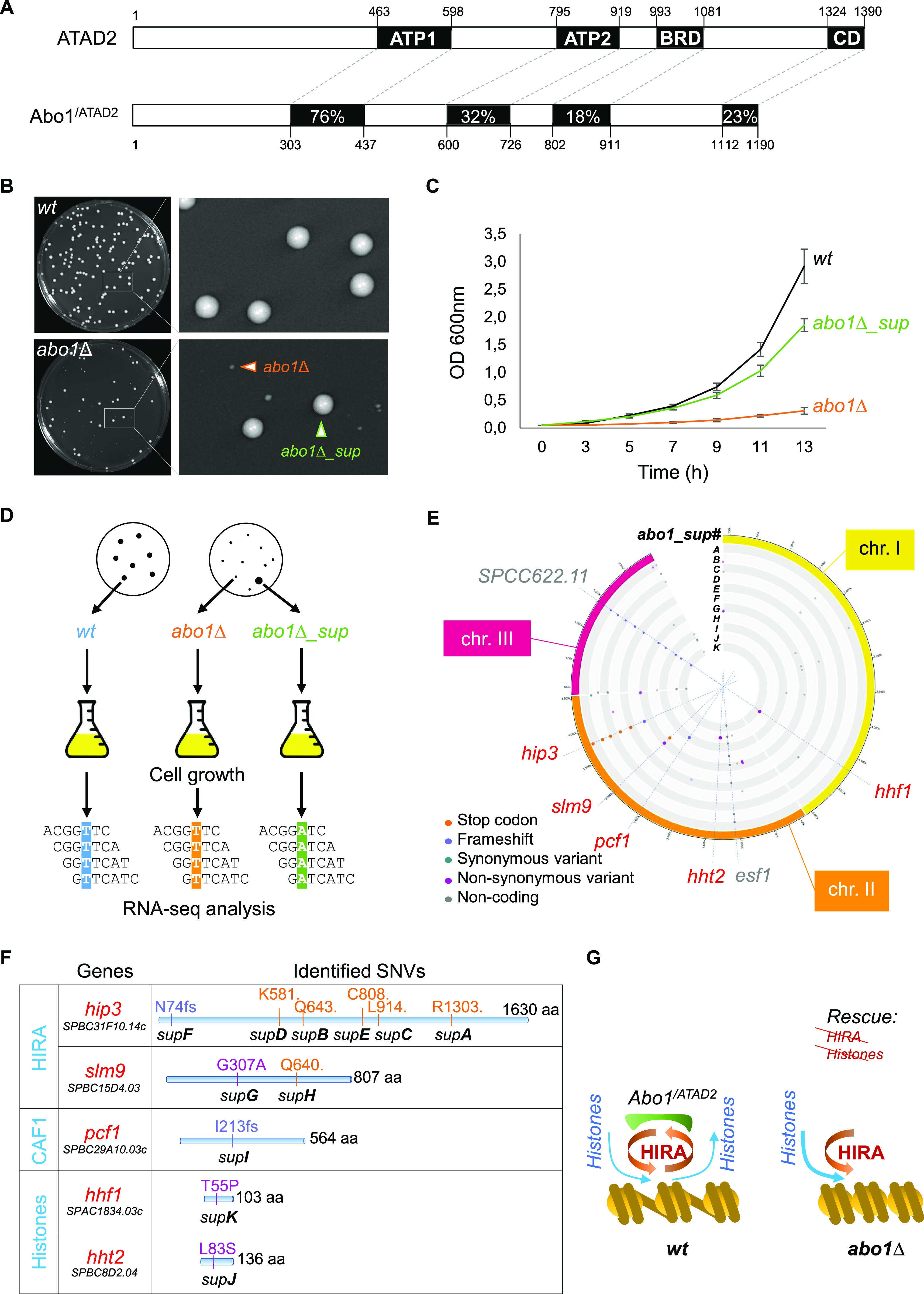
(A) Schematic representation of human ATAD2 and S. pombe Abo1 proteins. The percentage of amino acid sequence identity is indicated for the most conserved domains. ATP1 and ATP2, AAA+ ATPase domain 1 and 2; BRD, Bromo domain; CD, C-terminal domain. (B) Photographs of wild-type (wt, SPV 8) and abo1∆ (SPV 3789) colonies grown on solid medium for 4 d, with magnified views to highlight the co-existence of small size colonies (orange arrowhead) and large size colonies (green arrowhead, which correspond to suppressor clones) on the abo1∆ plate. (C) Growth curves of wt, abo1∆, and abo1∆ suppressor (abo1∆_sup) cells obtained from three biological replicates done with three different clones (SPV 3,789, 3,790, and 3,791) and grown in liquid medium over the indicated times. (D) Diagram of the screen conducted to identify mutations responsible for the suppression of abo1∆ cell growth’s defect by using RNA sequencing. Single colonies of wt, abo1∆ (with severe growth defect) and abo1∆_sup cells were picked to inoculate liquid cultures and total RNA was purified. Massive parallel sequencing from the purified total RNAs depleted from rRNAs was used to detect SNVs (for details see Table S1). Spotting assays of wild-type, abo1∆ and abo1∆_suppressor clones are shown in Fig S1. (E) Circos plot showing the SNVs identified in 11 independent isolates of abo1∆_sup (A to K) along the three S. pombe chromosomes (chr. I, II and III). The positions of all SNVs identified, either in coding or non-coding regions of the genome, are highlighted by dots colored according to the nature of the mutations, as indicated. Blue dashed lines highlight SNVs localizing within the coding sequence (cds) of subunits of histone chaperone complexes (hip3, slm9, and pcf1) or histones (hht2 and hhf1) genes, as well as two SNVs, in the cds of SPCC622.11 or esf1 genes, identified in a majority of abo1∆_sup isolates but that do not contribute to the suppression of abo1∆ cells growth defect. Spotting assays of backcrossed abo1∆_suppressor clones are shown in Fig S3. (F) Position and nature of the amino acid modifications induced by the SNVs found in the 11 abo1∆_sup isolates. The color code of the mutations is the same as in (E). fs: frameshift, dot: non-sense mutation. (G) Scheme showing the functional interplay between Abo1 and HIRA complex allowing correct loading of histones on chromatin in wild-type cells (left part of the panel), and the potential effect of a lack of Abo1 on HIRA’s function and histone loading (right part of the panel) in abo1∆ cells.
