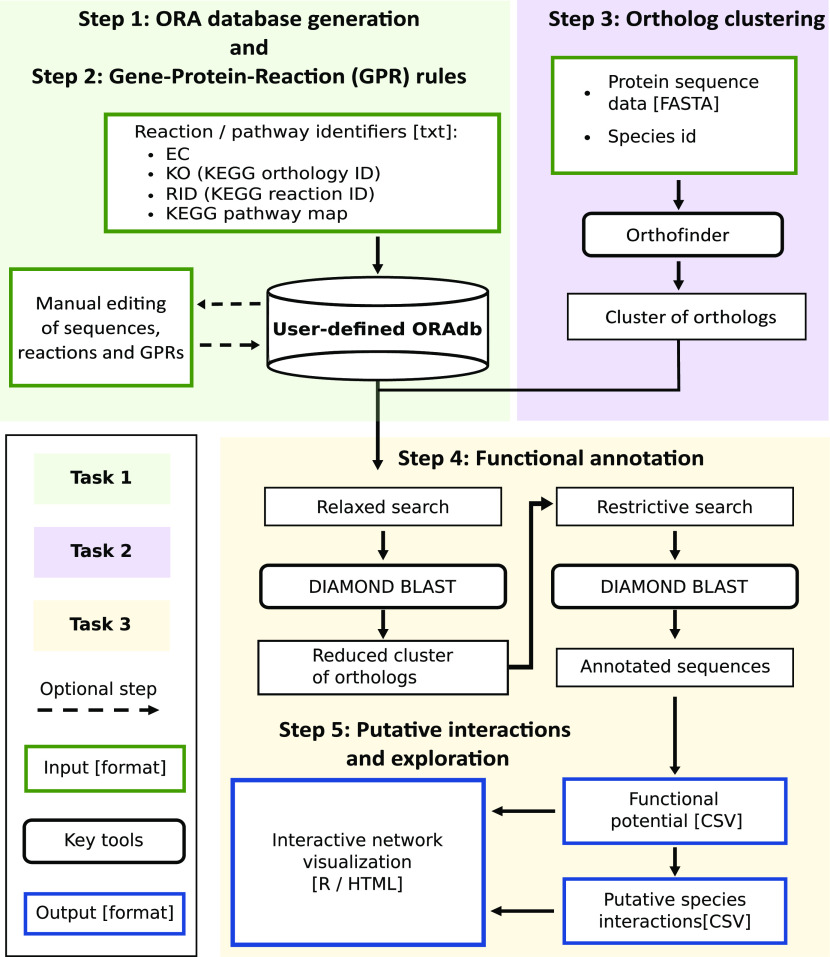
Figure 1. OrtSuite workflow.
OrtSuite takes a text file containing a list of identifiers for each reaction in the pathway of interest supplied by the user to retrieve all protein sequences from KEGG Orthology and are stored in ORAdb. Subsequently, the same list of identifiers is used to obtain the gene-protein-reaction (GPR) rules from KEGG modules (Task 1). Protein sequences from samples supplied by the user are clustered using OrthoFinder (Task 2). In Task 3, the functional annotation, identification of putative synergistic species interactions and graphical visualization of the network are performed. The functional annotation consists of a two-stage process (relaxed and restrictive search). Relaxed search performs sequence alignments between 50% of randomly selected sequences from each generated cluster. Clusters whose representative sequences share a minimum E-value of 0.001 to sequences in the reaction set(s) in ORAdb continue to the restrictive search. Here, all sequences from the cluster are aligned to all sequences in the corresponding reaction set(s) to which they had a hit (default E-value = 1 × 10−9). Next, the annotated sequences are further filtered to those with a bit score greater than 50 and are used to identify putative microbial interactions based on their functional potential. Constraints can also be added to reduce the search space of microbial interactions (e.g., subsets of reactions required to be performed by single species, transport-related reactions). In addition, an interactive network visualization of the results is produced and accessed via a HTML file.