Figure 1. Concepts and design of SARS-CoV-2 RNA biosensors.
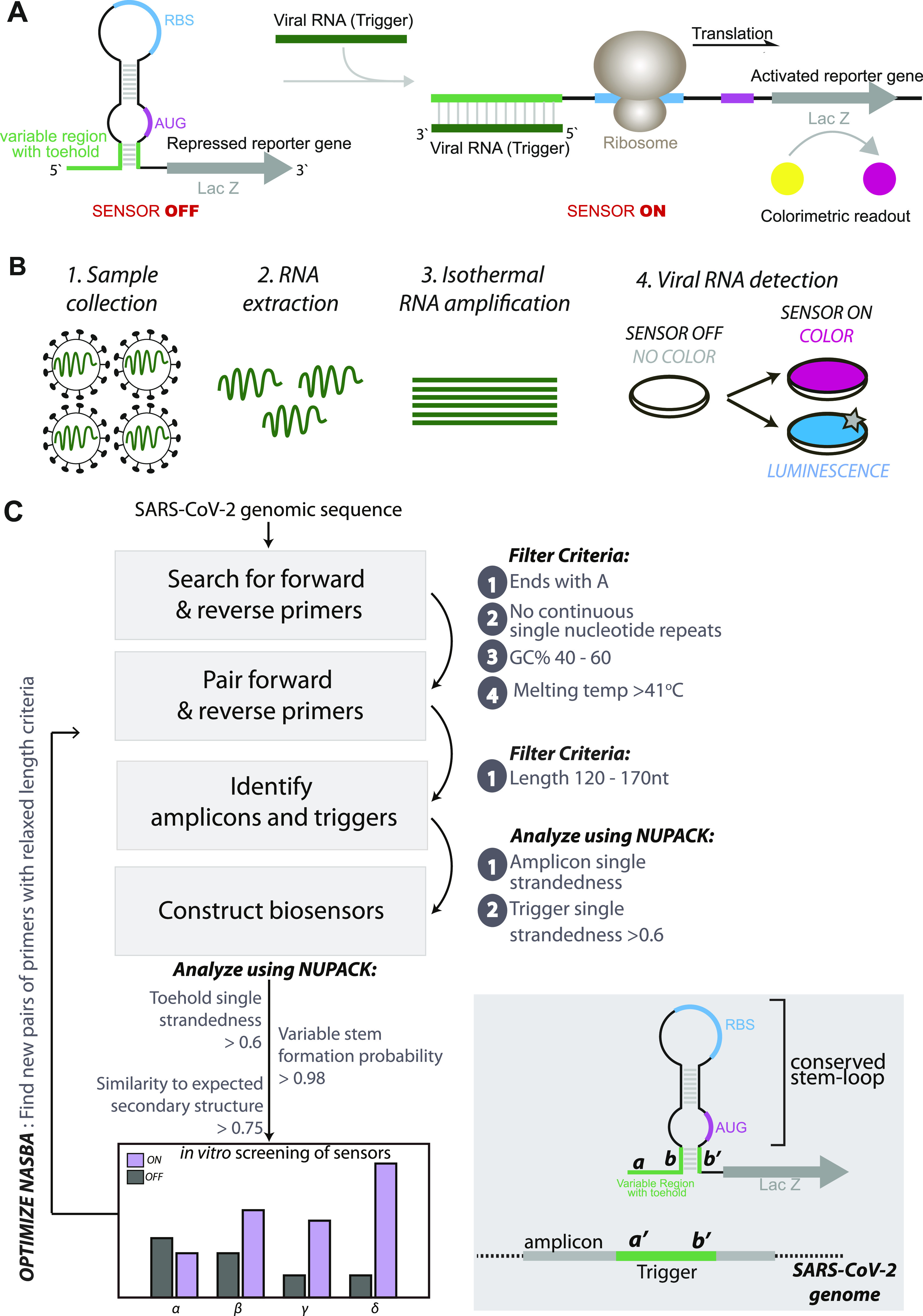
(A) Schematic of toehold switches. Toehold RNA switches consist of a central stem loop structure that harbors a ribosome binding site (RBS, blue) and a translation start site (AUG, pink) with a downstream reporter gene (such as lacZ, grey). A variable region with the toehold (green) is designed to specifically base-pair with a Trigger RNA (dark green). In the absence of Trigger RNA (left), the RBS and AUG are sequestered within the sensor structure and inaccessible to the ribosome. Presence of the Trigger RNA (right) induces intermolecular interactions between the toehold and the Trigger RNA, resulting in an alternate conformation wherein the RBS and AUG are accessible to the ribosome, enabling translation of the downstream LacZ enzyme. Production of LacZ is easily monitored with color, using a chromogenic substrate. The concept is modular and allows the use of alternate reporter genes and modes of detection. (B) Schematic showing our assay development pipeline. RNA extracted from viral particles is amplified isothermally using (NASBA) nucleic acid sequence–based amplification and detected with specifically designed toehold-based biosensors in an in vitro transcription-translation assay. The NASBA coupled in vitro transcription-translation assay leads to production of color that can be easily visualized by eye or with cell phone cameras or luminescence that can be quantified by luminometry. Our assay development pipeline focused on identifying targetable regions of the SARS-CoV-2 genome, design of specific biosensors, optimized primers for efficient NASBA, and overall sensitivity and response of the assay. (C) Flowchart showing the bioinformatic pipeline for primer design, and selection of biosensors. First, we searched for primers that would amplify fragments of the SARS-CoV-2 genome, with criteria as highlighted in the figure. Amplicons resulting from primer pairs were analyzed for potential Trigger regions. Amplicon and Trigger single strandedness were estimated. These Trigger regions were used to construct the biosensors, which were then analyzed for toehold single strandedness, stem probability and fidelity to the expected biosensor secondary structure. Illustration (bottom right) shows the elements of the biosensor and Trigger RNA in detail.
