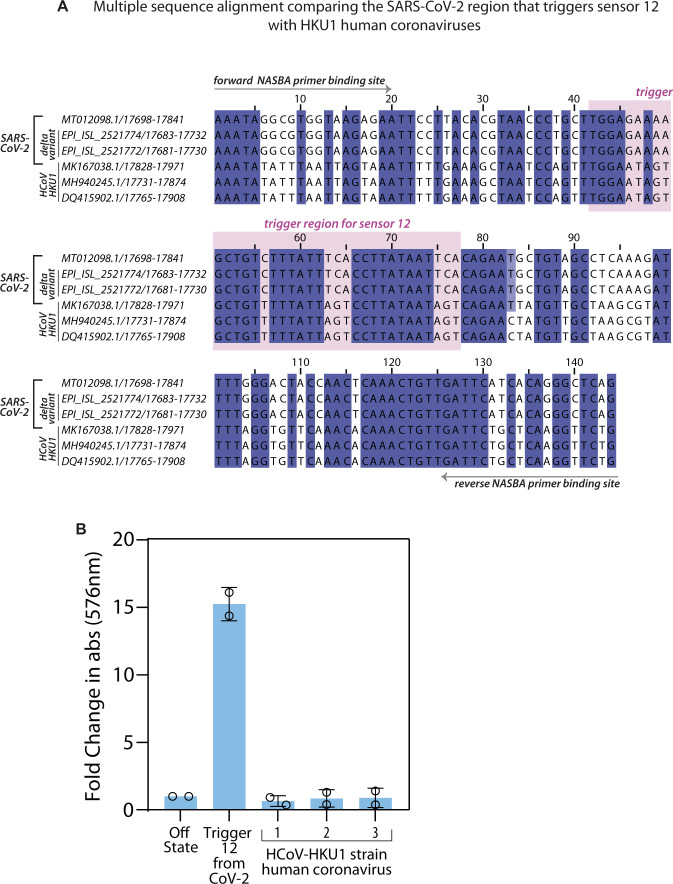
Figure S3. Selectivity of Sensor 12 for SARS-CoV-2 versus human corona viruses.
(A) Multiple sequence alignment (carried out using Clustal Omega) of a portion of the SARS-CoV-2 genome (MT012098.1), δ variant and HKU1 human corona viruses are shown. Region encompassing the cognate Trigger RNA that turns on Sensor 12, along with the binding sites for the forward and reverse NASBA primers (arrows) and Trigger 12 (pink box) are highlighted. (B) In vitro transcription–translation assays performed with sensor 12 were monitored and fold change in absorbance (576 nm) plotted. 1013 copies of the SARS-CoV-2 cognate Trigger RNA or equivalent RNA from HKU1 human corona viruses were added. Fold change is calculated relative to OFF state of sensor (sensor alone, no RNA added). Data are shown for two independent experiments (as indicated by the open circles). The error bars represent standard deviations. Human coronavirus RNAs did not show significant color compared with the off state.