Abstract
Genome sequences of eleven avian influenza virus (AIV) subtypes have been reported in swine populations from seven countries until August 2020. To unravel the transmission dynamics and spillover events of AIVs from avian reservoirs to swine, full-length hemagglutinin (HA) sequences of AIV subtypes (n = 11) reported from various avian species and swine were retrieved from the ‘Influenza Research Database’. Phylogenetic analysis identified closely related avian and swine AIV sequences suggesting potential spillover events from multiple domestic and wild avian species, including chicken, duck, pigeon, goose, quail, and aquatic birds to swine. Furthermore, N-linked glycosylation analysis of these closely related AIV sequences supported the possibility of multiple spillover events of highly pathogenic H5N1 and low pathogenic H9N2 viruses from various avian species to swine. The principal coordinate analysis further validated these findings for H5N1 and H9N2 viruses; however, spillover events of the other nine AIV subtypes were limited. Interestingly, the presence of potential mammalian adaptation markers, particularly in some of the swine H5N1, H7N9, and H9N2 viruses, suggested that these viruses may have already adapted in swine. The occurrence and circulation of these AIVs in swine, especially the H5N1 and H9N2 viruses with numerous spillover events from the avian reservoirs to swine, pose a significant threat in terms of their reassortment with endemic swine viruses or circulating human influenza viruses within the swine which may facilitate the emergence of a novel influenza virus strain with pandemic potential.
Keywords: Avian influenza virus, Avian to swine spillover, Phylogenetic analysis, N-linked glycosylation, Principal coordinate analysis, IAV adaptation, Virus evolution, Influenza pandemic
Introduction
Influenza A virus (IAV) is a member of the Orthomyxoviridae family with a broad avian and mammalian host range [34, 51, 54, 55, 57–59, 63]. Genomes of IAV comprise eight gene segments of a negative-sense single-stranded RNA [5]. The IAV virion particle has two surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA). The acquisition of subtle alterations in amino acid sequences within the HA protein triggers seasonal influenza epidemics [4, 49]. In contrast, the reassortments of different IAV subtypes co-infecting a host trigger the emergence of a novel IAV strain, which may be able to start a pandemic in an immunologically naive population [3, 4, 36, 38]. Due to the constant evolution of IAV, 18 HA and 11 NA subtypes have been reported infecting various avian and mammalian hosts [20, 27, 48].
The 1918 Spanish flu was the first and the deadliest of all the four influenza pandemics that occurred so far. One complete genome sequence of 1918 Spanish flu was recently obtained from a single victim buried under the permafrost in Alaska [40, 50], and four partial genome sequences were obtained from archival biopsies [40]. Analyses of these sequences and other contemporary IAV sequences determined that the 1918 H1N1 pandemic virus originated from an avian reservoir that was most likely transmitted to the human population shortly before the emergence of the Spanish flu pandemic [41]. More recently, the 2009 swine flu pandemic originated during March–May 2009 in the Mexican swine [14, 34]. The role of migratory birds and long-distance swine trades with human interaction were the primary factors responsible for the 2009 swine flu pandemic [14, 34].
While the first molecular evidence of interspecies transmission of AIV to swine was reported in Canada in 1999 when a low pathogenic H4N6 virus was detected in swine [22], followed by reports of H3N3 [23] in swine in Ontario in 2001, the majority of the reports of AIVs have been documented in Chinese swine: H9N2 in 2004 [8], H5N1, H7N9 [17] and H10N5 in 2008 [56], H4N1 in 2009 [19], H6N6 in 2010 [64], and H4N8 in 2011 [46]. Additionally, the genomes of H5N2 [30], H7N2 [29], and H9N2 viruses [12] were reported from South Korean swine in 2001, 2004, and 2008, respectively. The genomes of highly pathogenic H5N1 viruses were also reported from Indonesian swine during 2005–2007 [37]. Most recently, in 2018, the genome of an H5N2 virus was reported from Mexican swine [42].
The proteolytic cleavage of HA into HA1 and HA2 by the removal of an N-terminal signal sequence [33] facilitates the binding of IAV virion particles to the host's sialic acid (SA) receptors [26, 44, 52] and is an important step in IAV pathogenesis, as host receptor binding facilitates the internalization and membrane fusion of the IAV virion [45], triggering infection [10]. The HA protein then undergoes post-translational modification by a process termed 'N-linked glycosylation' [25] and a recent study reported the utility of using N-linked glycosylation patterns in HA protein to identify ecological spillover and adaptation of influenza viruses [24]. Addition or deletion of the glycosylation site(s) may impact IAV adaptation and infectivity in the host [9, 26]. In this context, we analysed full-length HA glycoprotein sequences of AIV subtypes (n = 11) reported in swine and various avian species globally to identify avian to swine spillover events and transmission dynamics.
Materials and methods
Acquisition of influenza virus sequences from the database
We downloaded all full-length HA nucleotide sequences of H3N3 (34 avian and two swine), H4N1 (13 avian and one swine), H4N6 (1394 avian and three swine), H4N8 (256 avian and one swine), H5N1 (3516 avian and 28 swine), H5N2 (1115 avian and four swine), H6N6 (419 avian and two swine), H7N2 (212 avian and one swine), H7N9 (746 avian and two swine), H9N2 (6301 avian and 22 swine), and H10N5 viruses (90 avian and one swine) available in the Influenza Research Database (https://www.fludb.org/) until 31 August 2020. The partial HA sequences were not included in the study.
Phylogenetic analysis
All full-length HA nucleotide sequences of AIV subtypes (n = 11) that have been reported in avian species and swine were used for phylogenetic analysis. The best substitution models for the phylogenetic analyses were identified using MEGA-X software [28]. Tamura-Nei (TN93) appeared to be the best model for the full-length nucleotide sequences under investigation. The full-length HA nucleotide sequences were then aligned using MUSCLE multiple sequence alignment in ‘Geneious Prime 2020.1.2’ software for constructing Neighbour-Joining phylogenetic trees. Each tree was created using the TN93 genetic distance model with 1000 bootstrap replications. We built the individual trees for each AIV subtype to determine the transmission patterns between avian species and swine. The neighbour-joining trees identified the most closely related sequences of avian and swine AIV subtypes to explore their N-linked glycosylation sites further.
Prediction of potential N-linked glycosylation sites in HA proteins
We identified the closely related AIV subtypes reported from avian species and swine (sharing the same nodes or clade) from the Neighbour-Joining trees for conducting N-linked glycosylation analysis. The N-linked glycosylation analysis was performed using NetNGlyc 1.0 web server, maintained by the Center for Biological Sequence analysis (http://www.cbs.dtu.dk/services/NetNGlyc/) at the Technical University of Denmark [15]. The NetNGlyc 1.0 web server examines the sequences in the context of Asn-Xaa-Ser/Thr sequons to predict the potential N-glycosylation sites in the protein sequence under query. The algorithm used in the NetNGlyc 1.0 web server obtains a 'jury agreement' for the nine artificial neural networks evaluated based on a threshold score of ˃0.5 [15, 21]. The NetNGlyc 1.0 server was chosen for the prediction of potential glycosylation sites because it has been reported to have a high accuracy of 76% [15, 21].
Principal coordinate analysis
The distance matrix for each subtype of AIV under investigation was generated using full-length HA nucleotide sequences in 'Geneious Prime 2020.1.2' software, providing a comma-separated values (CSV) file. The CSV file of the distance matrix was converted to the excel file using Microsoft Excel. Finally, the distance matrix was used for the principal coordinate analysis (PCA) using 'PAST: Paleontological Statistics Software Package for Education and Data Analysis' version 4.02 [16]. The PAST software package is a highly cited program that integrates spread-sheet-type distance matrix data with multivariate statistics, plotting and curve fitting, time series, and phylogenetic analysis [16].
Multiple sequence alignment
The full-length HA nucleotide sequences of swine H5N1 (n = 28) and H9N2 (n = 22) viruses were aligned using the MUSCLE algorithm in 'Geneious Prime 2020.1.2' software to assess their relatedness to ascertain the possibility of swine-to-swine transmission of these viruses.
Determination of mammalian adaptation markers
We identified previously reported potential mammalian adaptation markers in mature HA proteins of swine IAV subtypes (n = 11) using the Sequence Variation (SNP) tool available at the Influenza Research Database [65]. The Sequence Variation tool utilizes the HA subtype numbering conversion based on the cross-subtype HA numbering scheme proposed by Burke and Smith [6]. Additionally, the amino acid substitutions were manually verified using multiple or pairwise sequence alignment of swine IAV subtypes using the MUSCLE algorithm in ‘Geneious Prime 2021.2.2’ software.
Results
The present study utilized all the available full-length HA sequences of AIV subtypes (n = 11) in various avian species and swine populations to study avian to swine spillover events and transmission dynamics. The phylogenetics, N-linked glycosylation, and PCA analysis of AIV sequences under study identified multiple spillover events of highly pathogenic H5N1 and low pathogenic H9N2 viruses from various avian species to swine, while the spillover events of other nine AIVs were limited. We hereby presented the results of our analyses in a subtype-specific manner.
H5 subtypes (H5N1 and H5N2 viruses)
The highest number of full-length HA nucleotide sequences of AIVs in swine belonged to subtype H5N1 (n = 28), reported from China and Indonesia which along with HA sequences reported from various avian species globally (n = 3516) revealed the transmission dynamics and spillover events of H5N1 viruses from various avian species to swine (Fig. 1A). The phylogenetic analysis determined the closely related swine and avian H5N1 virus sequences suggesting multiple spillover events from avian species, including ducks, chicken, and wild birds to swine in Indonesia and China. The phylogenetic analysis of full-length HA nucleotide sequences of avian (n = 1115) and swine (n = 4) H5N2 viruses determined two independent events of avian to swine transmission of H5N2 viruses in Mexico and South Korea (Fig. 1A).
Fig. 1.
A Neighbour-joining tree of full-length HA nucleotide sequences of avian and swine H5N1 (n = 3544) and H5N2 viruses (n = 1119) suggested multiple spillover events from chicken, ducks and wild birds to swine. The possibility of swine-to-swine transmission of H5N1 viruses in Indonesia was also observed. The cyan nodes represented H5N1 virus transmission among avian species, while the dark cyan nodes represented H5N2 virus transmission among avian species. The red nodes represented H5N1 and H5N2 virus transmission from avian species to swine. The cyan and red dots at the nodes of the subtree represented > 90% bootstrap support. B Seven different glycosylation patterns of swine and closely related avian H5N1 viruses suggested multiple avian to swine transmissions of H5N1 viruses. The identical glycosylation patterns of swine and closely related avian H5N1 viruses represented the possibility of spillover within these groups. Two different patterns of N-linked glycosylation suggested independent introductions of H5N2 viruses in Mexican and Korean swine. C The PCA plot further confirmed multiple spillover events of H5N1 and H5N2 viruses in swine populations. The red dots represented H5N1 and H5N2 viruses in swine, while the cyan and dark cyan dots represented H5N1 and H5N2 viruses, respectively, in avian species reported worldwide. D The 100% amino acid as well as nucleotide sequence identities for a few swine H5N1 viruses (highlighted within two blue boxes) along with identical glycosylation sites (highlighted with red boxes) suggested the possibility of swine-to-swine transmission of these swine H5N1 viruses in Indonesia
The full-length amino acid sequences of closely related avian and swine H5N1 viruses identified from phylogenetics were further analysed for the N-linked glycosylation which determined the presence of seven patterns of glycosylation (Fig. 1B). The glycosylation sites at amino acid positions 26 (NNST), 27 (NSTE), 39 (NVTV), 302 (NSSM), 499/500 (NGTY), and 558/559 (NGSL) were conserved among all swine and closely related avian H5N1 viruses. The glycosylation sites at amino acid positions 88 (NVSE), 156 (NSSF or NPSF), 170 (NNTY), 179 (NYTN), 181 (NNTN), 209 (NPTT), and 289 (NCST) varied among the swine and closely related avian H5N1 viruses. Interestingly, a few Indonesian and Chinese swine H5N1 viruses had identical glycosylation sites which might be due to a common origin of these viruses given the long-distance migration network of avian H5N1 viruses in Southeast Asia [50]. Interestingly, the presence of seven different patterns of glycosylation in swine and closely related avian H5N1 viruses strongly supported the basis of phylogenetic clustering of these viruses.
The glycosylation sites at amino acid positions 26 (NNST), 27 (NSTE), 39 (NVTV), 181 (NNTN), 302 (NSSM), 496 (NGTY), and 555 (NGSL) were conserved among all swine and closely related avian H5N2 viruses, while glycosylation sites at amino acid positions 209 (NPTT) and 252 (NDSI) varied between Mexican and Korean swine and closely related avian H5N2 viruses (Fig. 1B). These findings supported the basis of the phylogenetic clustering of H5N2 viruses and confirmed the independent introductions of H5N2 viruses from avian species to swine in Mexico and South Korea.
The principal coordinate analysis (PCA) plot also supported the observations of phylogenetics and N-linked glycosylation and further added to the fact of the multiple spillover events of H5N1 viruses while suggesting two independent introductions of H5N2 viruses in swine (Fig. 1C). Due to limited number of available swine H5N2 virus sequences, the swine-to-swine transmission of H5N2 virus could not be ascertained. To our surprise, the multiple sequence alignment of HA nucleotide sequences of swine H5N1 viruses (n = 28) identified seven Indonesian swine with 100% nucleotide sequence identity, belonged to H5 clades 2.1.3 and 2.1.3.3 (Fig. 1D). Interestingly, these H5N1 viruses were reported from the same swine farm in Indonesia [37] which suggested a strong possibility of swine-to-swine transmission of H5N1 viruses.
H7 subtypes (H7N2 and H7N9 viruses)
The Neighbour-joining phylogenetic tree of full-length HA nucleotide sequences of avian H7N2 (n = 212) and swine H7N2 (n = 1) viruses determined wild bird to swine transmission of the H7N2 virus in South Korea (Fig. 2A). Two full-length HA gene sequences of highly pathogenic H7N9 viruses were reported from Chinese swine in 2017. The Neighbour-joining tree of these two Chinese swine H7N9 viruses with 746 avian H7N9 viruses reported worldwide (Fig. 2A) shows chicken to swine transmission of H7N9 viruses.
Fig. 2.
A Neighbour-joining tree of full-length HA nucleotide sequences of 212 avian and one swine H7N2 viruses represented wild bird to swine transmission of the H7N2 virus in South Korea. Neighbour-joining tree of 748 full-length HA nucleotide sequences of swine and avian H7N9 viruses determined chicken to swine spillover of H7N9 viruses in China. A subtree highlighted the circulation of H7N9 viruses between chicken and swine in China. The blue and red dots at the nodes of the subtree represented > 90% bootstrap support. B The identical glycosylation pattern in HA proteins of H7N2 viruses obtained from multiple wild bird species and ducks suggested a broad circulation of H7N2 viruses in avian species in Southeast Asia with one spillover event to swine. The identical N-linked glycosylation patterns shared by swine and closely related avian (chicken) H7N9 viruses supported the findings of phylogenetics suggesting chicken to swine spillover of H7N9 virus in China. C PCA plot represented the global transmission of avian H7N2 viruses with one spillover event to swine and two avian to swine spillover events of H7N9 viruses. The cyan and dark cyan dots represented the circulation of H7N2 and H7N9 viruses, respectively, among avian reservoirs worldwide, while the red dots suggested the occurrence of swine H7N2 and H7N9 viruses
The H7N2 viruses obtained from swine and phylogenetically closely related ducks and wild birds had identical glycosylation sites (Fig. 2B) which suggested a broad circulation of H7N2 viruses in avian species in Southeast Asia. The PCA plot depicted the global pattern of avian and swine H7N2 virus transmission (Fig. 2C). The HA nucleotide sequences of H7N9 viruses reported from Chinese swine were not identical which ruled out the possibility of swine-to-swine transmission of H7N9 viruses and therefore suggested two separate spillover events from chicken to swine. The presence of identical glycosylation sites in swine and closely related chicken H7N9 viruses further supported the phylogenetic analysis suggesting chicken to swine spillover of H7N9 viruses in China (Fig. 2B). Two overlapping red dots in the PCA plot suggested that the Chinese swine may have acquired the H7N9 virus either from the same poultry flock or a common origin (Fig. 2C).
H9N2 virus
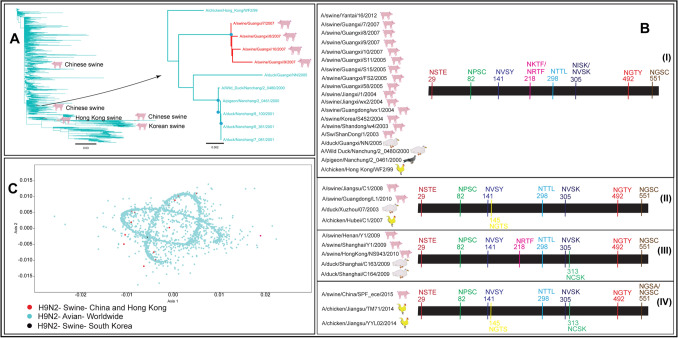
Total 6323 full-length HA gene sequences of low pathogenic H9N2 viruses have been reported from swine (n = 22) and avian species (n = 6301) worldwide. Neighbour-joining tree suggested multiple events of avian to swine transmission of H9N2 viruses in China, Hong Kong, and South Korea (Fig. 3A).
Fig. 3.
A Neighbour-joining tree of 6323 full-length HA nucleotide sequences of avian and swine H9N2 viruses determined multiple spillover events from avian reservoirs to swine. The red nodes represented avian to swine transmission of H9N2 viruses in China and Hong Kong, while black nodes represented avian to swine spillover in South Korea. The cyan nodes represented the circulation of H9N2 viruses among avian species globally. The blue dots at the nodes in the subtree represented > 90% bootstrap support. B Four patterns of glycosylation sites in swine and avian H9N2 viruses suggested multiple introductions and evolutionary trajectories of H9N2 viruses in swine in Southeast Asia. C The PCA plot represented multiple spillover events of H9N2 viruses in swine
The phylogenetic analysis was supported by the N-linked glycosylation patterns which suggested at least four evolution trajectories of H9N2 viruses in Chinese swine (Fig. 3B). Additionally, the phylogenetics and glycosylation patterns suggested only one spillover event of H9N2 viruses in each of Hong Kong and South Korean swine populations. Interestingly, identical glycosylation patterns among Chinese, Hong Kong and South Korean swine populations indicated a common origin of H9N2 viruses which might be due to either through long-distance bird migration or swine trade in Southeast Asia. The maximum number of swine H9N2 viruses were reported from China which suggested a widespread circulation of H9N2 viruses in avian species in China and their frequent interactions with swine.
The glycosylation sites at amino acid positions 29 (NSTE), 82 (NPSC), 141 (NVSY), 298 (NTTL), 305 (NVSK), 492 (NGTY) and 551 (NGSC or NGSA) were conserved, while glycosylation sites at amino acid positions 145 (NGTS), 218 (NRTF or NKTF), and 313 (NCSK) were unstable among swine and avian H9N2 viruses. The PCA plot supported the phylogenetics and N-linked glycosylation analyses and depicted multiple spillover events of H9N2 viruses in swine (Fig. 3C). Most of the swine H9N2 viruses except two differed in nucleotide sequences among each other suggesting a low possibility of swine-to-swine transmission.
H3N3 virus
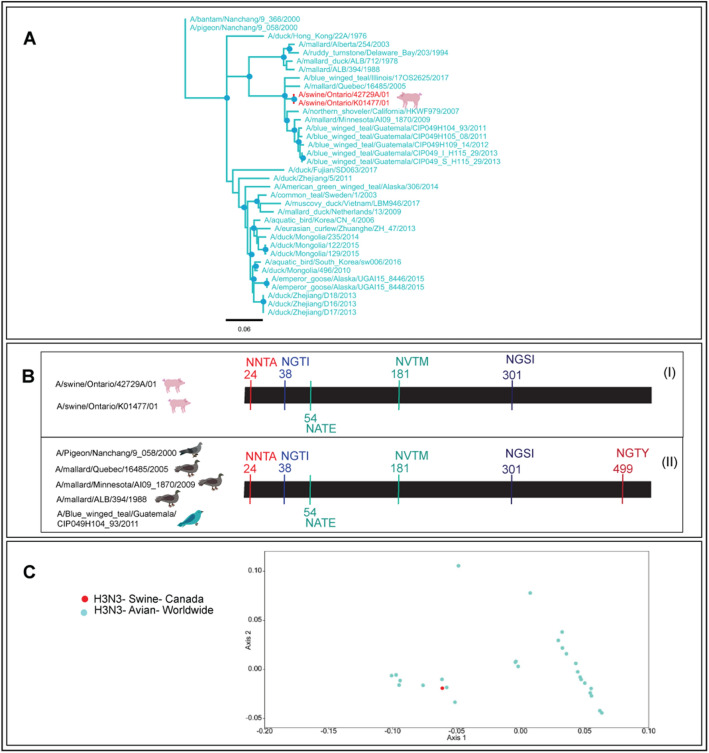
Thirty-six full-length HA gene sequences of avian and swine H3N3 viruses including two sequences from Ontario based swine were analysed. The Neighbour-joining tree suggested that the transmission of H3N3 virus to Ontario based swine may have occurred from wild birds (Fig. 4A).
Fig. 4.
A Neighbour-joining tree of 36 full-length HA nucleotide sequences of avian and swine H3N3 viruses determined the wild bird to swine transmission in Ontario. The blue dots at the nodes represented > 90% bootstrap support. B The loss of glycosylation at amino acid residue 499 (NGTY) appeared to have transmitted H3N3 virus to swine; however, it should be noted that the availability of a limited number of avian and swine H3N3 virus sequences restricted our ability to infer avian to swine spillover. C The PCA plot illustrated the occurrence of swine and avian H3N3 viruses globally
The N-linked glycosylation analysis suggested that deglycosylation at amino acid position 499 (NGTY) may have transmitted the H3N3 virus to the swine (Fig. 4B). The PCA plot determined an overlapping red dot for swine H3N3 viruses in Ontario (Fig. 4C) which suggested a common avian origin. It should be noted that the availability of limited sequence data for H3N3 virus HA genes made it challenging for us to identify the potential spillover event from avian to swine. The lack of further reports of H3N3 virus in swine suggested that the H3N3 virus might have got introduced to Ontario swine through a sporadic transmission from an avian reservoir which did not spread further and hence was never detected again.
H4 subtypes (H4N1, H4N6, and H4N8 viruses)
The phylogenetic analysis of full-length HA gene sequences of H4N1 (13 avian and one swine), H4N6 (1397 avian and three swine), and H4N8 (256 avian and one swine) viruses determined duck to swine spillover of H4N1 and H4N8 viruses in China, and mallard to swine transmission of H4N6 viruses in Canada and the United States (Fig. 5A).
Fig. 5.
A Neighbour-joining tree of avian and swine H4N1, H4N6, and H4N8 virus HA nucleotide sequences depicted duck to swine transmission of H4N1 and H4N8 viruses in China and mallard to swine transmission of H4N6 viruses in North America. The blue and red dots at the nodes in the subtree represented > 90% bootstrap support. B The identical N-linked glycosylation patterns of swine and duck H4N1 and H4N8 viruses supported the phylogenetic clustering. The glycosylation at amino acid position 14 (NSSQ) appeared crucial for duck to swine spillover of H4N1 virus. The identical glycosylation sites of swine and closely related mallard H4N6 viruses suggested mallard to swine transmission of H4N6 virus in North America. C The PCA plot represented the occurrence of swine and avian H4 subtypes which supported the phylogenetic analyses
The presence of identical glycosylation sites in HA proteins of Chinese swine and Hong Kong ducks (Fig. 5B) strengthened the outcome of phylogenetic analysis. The glycosylation at amino acid residue 14 (NSSQ) appeared crucial for duck to swine transmission of the H4N1 virus. The PCA plot supported the phylogenetic and glycosylation analyses and grouped the swine H4N1 virus along with the Asian and European avian H4N1 viruses. It was interesting to note that H4N1 viruses reported from Chinese swine and Hong Kong duck were not contemporary which suggested that the H4N1 virus may have been in circulation in avian reservoirs in Southeast Asia at a low frequency and hence remained undetected. The presence of identical glycosylation sites in closely related HA proteins of swine and avian (mallard) H4N6 viruses (Fig. 5B) suggested the circulation and long-distance dissemination of avian H4N6 viruses in North American mallards. These observations were further supported by the PCA plot which depicted independent spillover events of H4N6 viruses in North American swine (Fig. 5C). Presence of the identical glycosylation sites in H4N8 viruses obtained from swine and ducks in China as well as ducks in South Africa (Fig. 5B) along with shared phylogenetic clustering suggested a long-distance intercontinental dissemination of H4N8 viruses. These findings were concerning regarding the existing risk of avian to swine transmission of H4N8 viruses in South Africa. The global transmission of H4N8 viruses in avian species and swine was further depicted using a PCA plot (Fig. 5C).
H6N6 virus
The neighbour-joining tree of full-length HA gene sequences of swine H6N6 (n = 2) and avian H6N6 (n = 419) viruses suggested two separate events of avian to swine transmission in China (Fig. 6A). One event suggested a pigeon to swine transmission of H6N6 virus in China, while the other event suggested a duck to swine transmission. The identical glycosylation pattern (Fig. 6B) suggested a common origin and widespread occurrence of H6N6 viruses in Chinese avian species. The PCA analysis confirmed two independent transmission events of H6N6 viruses in Chinese swine (Fig. 6C).
Fig. 6.
A Neighbour-joining tree of 421 full-length HA nucleotide sequences of swine and avian H6N6 viruses represented two separate events of avian to swine transmission in China highlighted with red nodes. The blue dots at the nodes in the subtree represented > 90% bootstrap support. B The identical glycosylation patterns of pigeon, duck, and swine H6N6 viruses in China suggested a broad circulation of H6N6 viruses in avian species and their sporadic spillover to swine. C The PCA plot confirmed the occurrence of two independent spillover events of H6N6 viruses in Chinese swine
H10N5 virus
The neighbour-joining phylogenetic tree of 90 avian and one swine full-length HA sequence of H10N5 viruses suggested chicken to swine transmission of the H10N5 virus in China. It was interesting to note that these closely related chicken and swine H10N5 virus sequences were not contemporary, suggesting a circulation of H10N5 viruses in various domestic and wild avian species in China and Southeast Asia (Fig. 7A) which may be circulating at a low frequency and hence remained undetected. The identical glycosylation patterns of closely related swine and various avian H10N5 virus HA proteins suggested a common origin of H10N5 viruses in avian species in China (Fig. 7B). The PCA plot depicted the transmission of H10N5 viruses in swine and avian species (Fig. 7C). The H10N5 virus appeared to be in circulation in avian species in China and the Southeast Asia at a low frequency.
Fig. 7.
A Neighbour-joining tree of 91 full-length HA nucleotide sequences of H10N5 viruses reported from avian species and swine determined one spillover event from chicken to swine in China. The closely related chicken and swine H10N5 virus sequences were not contemporary suggesting a circulation of these viruses in avian species at a low frequency in the region. The blue dots at the nodes in the subtree represented > 90% bootstrap support. B The identical glycosylation pattern of chicken and duck H10N5 viruses suggested a common origin of circulating H10N5 viruses in China with one spillover event to swine. C The PCA plot represented the global transmission pattern of avian and swine H10N5 viruses
In summary, the HA gene sequences of eleven AIV subtypes, including highly pathogenic H5N1 and H7N9, and low pathogenic H3N3, H4N1, H4N6, H4N8, H5N2, H6N6, H7N2, H9N2, and H10N5 viruses, have been reported in swine until 31 August 2020. The neighbour-joining phylogenetic trees followed by N-linked glycosylation and PCA analyses determined multiple spillover events for highly pathogenic H5N1 viruses in Chinese and Indonesian swine. There were also numerous spillover events for low pathogenic H9N2 viruses in Chinese swine; however, a limited spillover occurred for other AIV subtypes.
Adaptation of avian-origin IAVs in swine
Several amino acid substitutions in HA glycoproteins of IAV subtypes have been reported to determine their preference of host receptor binding, which as a result, may facilitate interspecies transmission and adaptation of avian IAVs in a mammalian host, including swine. Therefore, we analysed potential mammalian adaptation markers in mature HA proteins of swine IAV subtypes (Fig. 8).
Fig. 8.
A–G Schematic representation of potential mammalian adaptation markers (H3 numbering) [6] in mature HA proteins of swine IAVs. The analysis was conducted using the Sequence Variation (SNP) tool of the Influenza Research Database [65]. A key amino acid substitution for mammalian adaptation, Q226L, was observed in swine H4N6, H7N9 and some of the H9N2 viruses, while another potential mammalian adaptation marker, T160A, was present in several swine H5N1, H5N2, H7N2, H7N9 and H9N2 viruses. A potential swine adaptation marker, D225G [32], was present in all of the swine H9N2 viruses
The presence of 226L and 228S (H3 numbering) in the HA protein of H4N6 viruses suggested that swine H4N6 viruses have already adapted for mammalian transmission, since these are the key markers associated with increased α-2,6- SA receptor binding ability facilitating mammalian adaptation and transmission [6]. The other swine H4 subtypes, including H4N1 and H4N8 viruses, had 226Q and 228G, which prefer α-2,3-SA receptor binding and therefore do not appear to be adapted in swine for efficient mammalian transmission. Intriguingly, swine H5N1 viruses, despite lacking the above key adaptation markers, have acquired other potential markers, including S137A, I155T, N158D, T160A, T192I, and K193R. The T160A and T192I substitutions were also observed in some of the swine H5N2 viruses. Since these amino acid substitutions prefer α-2,6-SA receptor binding, the probability of swine-to-swine transmission of these swine H5 viruses remains feasible.
Interestingly, the T160A substitution was also observed in other swine viruses, including H7N2, H7N9, and H9N2. From these observations, it appears that the Thr to Ala substitution at amino acid position 160 would be essential for swine adaptation of avian-origin H5, H7, and H9 viruses; however, it warrants further investigation using swine model. Interestingly, swine H7N9 and some of the swine H9N2 viruses also had a critical mammalian adaptation marker, Q226L. In addition to this, all swine H9N2 viruses (n = 22) had the D225G substitution, which in a previous study was suggested to be required for swine adaptation of H9N2 viruses [32]. One of the two swine H6N6 viruses has acquired A222V and G228S substitutions, which prefer α-2,6-SA receptor binding while retaining 226Q, which prefers α-2,3-SA receptor binding. The other swine viruses, including H3N3 (n = 2) and H10N5 (n = 1), did not appear to carry potential mammalian adaptation markers in their HA proteins.
Overall, in our analysis, the presence of Q226L and G228S substitutions in swine H4N6 viruses suggested the possibility of their adaptation in swine. The presence of T160A and other potential substitutions in some of the swine H5N1 and H5N2 viruses suggested the possibility of their adaptation in swine. The presence of T160A and Q226L substitutions in swine H7N9 and some of the swine H9N2 viruses along with D225G substitution in swine H9N2 viruses suggested a strong possibility of their adaptation in swine.
Discussion
In a recent systematic review, we reported the prevalence of multiple AIV subtypes in swine populations [7] which warranted further investigation into the transmission dynamics and possible spillover events of these viruses. In the present study, we used phylogenetic analysis followed by N-linked glycosylation and PCA analyses to unravel the events of avian to swine transmission of various AIV subtypes. Intriguingly, highly pathogenic H5N1 viruses had multiple sporadic transmissions from avian to swine in China and Indonesia. Similarly, numerous sporadic events of avian to swine transmission of low pathogenic H9N2 viruses were also identified in China, with limited avian to swine transmission observed for the other AIV subtypes. Since China is considered an epicentre of avian influenza viruses, multiple reports of avian H5N1 and H9N2 viruses from China were not surprising. However, the occurrence of numerous independent events of avian to swine transmission of H5N1 viruses in Indonesian swine suggested a broad intracontinental circulation of highly pathogenic H5N1 viruses in Southeast Asia. Mine et al. [35] reported that the migratory flyways of wild birds for inter-and intra-continental dissemination of IAVs appear to be responsible for higher H5N1 and H9N2 virus circulation in Southeast Asia [35]. To our surprise, we found two groups of identical HA nucleotide sequences of H5N1 viruses obtained from seven Indonesian swine, which strongly suggested the possibility of swine-to-swine transmission belonging to two H5 clades (2.1.3 and 2.1.3.3). Since IAV RNA polymerases have a low fidelity with a substitution rate of 2 × 10–5 per nucleotide [39, 43, 53], the probability of unrelated H5N1 viruses having identical nucleotide sequences is low.
We also found that those swine and avian IAVs that were closely related phylogenetically, appeared to have an additional glycosylation site in the HA protein. An exception was seen for H3N3 viruses, where the loss of a glycosylation site at position 499 (NGTY) appeared to have triggered the transmission from avian to swine. The highest variation in the pattern of the glycosylation sites was observed in the highly pathogenic H5N1 viruses. Seven different glycosylation patterns were observed for H5N1 viruses which suggested multiple sporadic transmission events of H5N1 viruses from avian reservoirs to swine. Similarly, the glycosylation sites at positions 145 (NGTS), 218 (NRTF or NKTF), and 313 (NCSK) were unstable in swine and avian H9N2 viruses, having four patterns of glycosylation which suggested multiple origins of swine H9N2 viruses as well.
The N-linked glycosylation deploys multiple functions to promote IAV fitness inside the host cell and hence plays a vital role in IAV evolution and adaptation in the host. Kim et al. [24] suggested that analyses of N-linked glycosylation patterns of HA protein may provide signatures for the identification of ecological spillover and adaptation of AIVs in swine [24]. Interestingly, the occurrence of identical glycosylation patterns in HA proteins of H4N8 viruses in Chinese ducks and swine and South African ducks and wild birds suggested a broad intercontinental bird migration pattern with an existing risk of avian to swine spillover of the H4N8 virus in South Africa.
While there have been reports of sporadic cases of highly pathogenic H5N1 virus transmission directly from avian (poultry) to human within family clusters in Indonesia in 2006 and Turkey during December 2015 to January 2016 [61], there has been limited human-to-human transmission. As a result, these infections have not yet been able to trigger an epidemic. However, the adaptation of highly pathogenic H5N1 virus in swine, either through mutation(s) or selective pressure [41], or swine-swine transmission, may facilitate a broader mammalian or human-to-human transmission.
The amino acid substitutions that confer increased binding affinity for the α-2,6-SA receptors have been identified as key mammalian adaptation markers that could facilitate mammalian or human-to-human transmission. We observed that swine H4N6 viruses had two key mammalian adaptation markers, 226L and 228S (H3 numbering) in the HA protein. A previous study reported that strains with the 226L mutation acquired binding affinity for α-2,6-SA receptors and induced higher infectivity in primary swine respiratory epithelial cells compared to H4N6 virus with 226Q [2]. Multiple passages of wildtype and mutant H4N6 viruses in swine respiratory epithelial cells revealed that the Q226L substitution was primarily related to swine adaptation of the H4N6 virus [2]. Interestingly, G228S did not appear to be essential for the adaptation of H4N6 virus in swine respiratory epithelial cells [2]. Despite the presence of 226L and 228S in swine H4N6 viruses, they had a limited replication in swine upper respiratory tract [1]. The presence of lesions and the detection of H4N6 virus in the lungs of experimentally inoculated piglets were reported with a limited transmissibility [1] which suggested a low risk of H4N6 virus transmission in the field setting.
In the present study, six amino acid substitutions: S137A, I155T, N158D, T160A, T192I, and K193R (H3 numbering), were observed in some of the swine H5N1 viruses which have been previously associated with enhanced affinity for α-2,6-SA receptor binding [13, 18, 59, 62] which suggests that these six amino acid substitutions could have facilitated the adaptation of H5N1 virus in swine. Additionally, T160A and N158D substitutions were found to be associated with the loss of glycosylation in the HA protein and H5N1 transmission in guinea pigs and ferrets [13, 18]. It was interesting to note that swine H5N1 viruses did not show any of the key adaptation markers, Q226L and G228S. Intriguingly, in our study, we observed dual substitutions, T160A and K193R, in six swine H5N1 viruses, which were indicative of their potential for increased α-2,6-SA receptor binding and adaptation in swine.
Lipatov et al. [17] showed that avian H5N1 virus in swine could be detected in nasal swabs of intranasally inoculated piglets up to 5 days post inoculation (dpi). While they did not cause any behavioural and food consumption changes in the swine [31], the infection was limited to the respiratory tract, mainly the lungs causing alveolitis and bronchiolitis [31]. Similarly, the intranasal inoculation of swine with a low pathogenic avian H5N2 virus also resulted in subclinical infections; however, the virus could be detected in nasal swabs up to 6 dpi [11] with virus more frequently isolated from the swine upper respiratory tract [11]. Taken together, these findings support the possibility of replication of H5N1 and H5N2 viruses in swine. It is possible that our observation of the T160A and T192I substitutions in some of the swine H5N2 viruses could be related to its replication in swine which warrants further investigation.
Further, we observed A222V and G228S substitutions in one of the swine H6N6 viruses. Interestingly, G228S but not the A222V substitution was previously identified to be associated with increased binding affinity to guinea pig erythrocytes [47]. While the authors showed that virus could be detected in nasal wash fluids of inoculated ferrets up to 5 dpi, the infection was subclinical. Interestingly, only one of the three ferrets that were in a direct contact with the virus inoculated ferret had the seroconversion; however, no virus shedding could be detected in direct contact ferrets suggesting a limited transmissibility of the H6N6 virus with A222V and G228S dual substitutions [47]. This suggested that swine H6N6 virus with A222V and G228S substitutions may have a limited transmissibility in swine and is yet to be adapted in swine to acquire critical substitutions for efficient mammalian transmission.
It was interesting to note that the four HA mutations reported by Xu et al. [60] (G63C, P65L, R66L, and M67I) acquired after the serial passage of an avian H7N9 virus in swine were different from the T160A and Q226L mutations observed in the naturally infected swine H7N9 viruses analysed in the present study. The passaged viruses could be detected in nasal and throat swabs of inoculated swine up to four dpi with focal mild interstitial pneumonia and lung infection at day four; however, the infection was always subclinical [60]. Intriguingly, despite the absence of the key Q226L substitution, the avian H7N9 virus was reported to rapidly adapt in the swine [60], which suggested that various amino acid substitutions, including T160A and Q226L, observed in the present study, may facilitate the adaptation of avian H7N9 virus in swine.
Neumann et al. [32] reported that avian H9N2 viruses with the 226L mutation were able to replicate in the upper respiratory tract of swine, with virus shedding in nasal swabs up to eight dpi [32]. They also reported an additional mutation, D225G, that was observed after four virus passages in swine. The passaged H9N2 virus with dual mutations, 225G and 226L, replicated and transmitted more efficiently in swine compared to the parental H9N2 virus with 225D and 226L [32]. Also, the H9N2 virus became contact transmissible after it acquired 225G, although the infections were always subclinical [32]. In our study, we observed that D225G was present in all swine H9N2 viruses (n = 22), while dual mutations (D225G + Q226L) were present in seven naturally infected swine. This suggested that swine H9N2 virus would be able to replicate and transmit between swine via contact in a natural setting. In addition, T160A was observed in most of the swine H9N2 viruses (n = 19). This mutation was also consistently present in several H5 and H7 viruses, suggesting that it had facilitated swine adaptation of these avian viruses.
Overall, the present study provided evidence of multiple spillover events of avian-origin H5N1 and H9N2 viruses in swine populations. The occurrence of certain putative mammalian adaptation markers, especially in swine H5N1, H7N9, and H9N2 viruses, suggested that these viruses have already adapted in swine, with supporting reports of these strains being able to replicate in the upper respiratory tract of swine. The replication and adaptation of these avian viruses in swine may facilitate the emergence of a pandemic influenza virus strain through reassortment with endemic swine viruses and circulating human IAVs in swine.
Conclusion
The interaction of swine with domestic and wild avian species may facilitate the interspecies transmission of avian IAVs to swine. Avian to swine spillover of AIVs, especially H5N1, H7N9, and H9N2 viruses, which have acquired some of the amino acid substitutions associated with mammalian adaptation, poses a considerable threat regarding the potential for the emergence of a pandemic influenza virus strain. Because the largest number of available full-length HA nucleotide sequences from swine belonged to either highly pathogenic H5N1 (n = 28) or low pathogenic H9N2 viruses (n = 22), this allowed us to use these two subtypes to understand their circulation and adaptation in swine. Numerous spillover events of H5N1 and H9N2 viruses from birds to swine suggest a high frequency of these viruses in avian reservoirs. In light of these risks, we recommend that swine farmers implement adequate biosecurity measures to minimize avian–swine interactions and that active IAV surveillance to monitor virus evolution in swine populations is imperative in order to combat the emergence of an influenza strain with pandemic potential.
Acknowledgements
The authors acknowledge the College of Health Sciences at the University of KwaZulu-Natal, Durban, for granting a three-year research scholarship to Ravendra P. Chauhan. The authors thank Dr. Apoorv Tiwari of G.B. Pant University of Agriculture and Technology, Pantnagar, India, for suggesting the software for PCA analysis. The authors are immensely thankful to both the reviewers and the Editor for providing useful suggestions which helped the authors to improve the manuscript.
Author contributions
Conceptualization: RPC and MLG; Methodology: RPC; Formal analysis and investigation: RPC; Writing—original draft preparation: RPC; Writing—review and editing: RPC and MLG; Critical Revision: RPC and MLG; Supervision: MLG. Both authors have read the present version of the manuscript and agreed to the publication.
Funding
This research received no external funding.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The influenza virus sequences analysed in this study were downloaded from the Influenza Research Database (https://www.fludb.org/). This research is part of a research project which has obtained full approval from the Animal Research Ethics Committee (AREC) of the University of KwaZulu-Natal, Durban; Reference: AREC/041/019D. Additionally, the authors have permission in terms of section 20 of the Animal Diseases Act, 1984 (Act No. 35 of 1984) from the Department of Agriculture, Land Reform and Rural Development (DALRRD), South Africa; Reference: 12/11/1/5/4 (1425).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abente EJ, Gauger PC, Walia RR, Rajao DS, Zhang J, Harmon KM, Killian ML, Vincent AL. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology. 2017;511:56–65. doi: 10.1016/j.virol.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman AC, Busch MG, Karasin AI, Bovin N, Olsen CW. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for alpha2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J Virol. 2008;82:8204–8209. doi: 10.1128/JVI.00718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshe RB. The origins of pandemic influenza–lessons from the 1918 virus. N Engl J Med. 2005;353:2209–2211. doi: 10.1056/NEJMp058281. [DOI] [PubMed] [Google Scholar]
- 4.Bertram S, Glowacka I, Steffen I, Kühl A, Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010;20:298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke DF, Smith DJ. A recommended numbering scheme for influenza A HA subtypes. PLoS ONE. 2014;9:e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan RP, Gordon ML. A systematic review analyzing the prevalence and circulation of influenza viruses in swine population worldwide. Pathogens. 2020;9:355. doi: 10.3390/pathogens9050355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong YL, Wang CF, Yan CM, Peng JS, Jiang ZL, Liu JH. Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes. 2008;36:461–469. doi: 10.1007/s11262-008-0227-z. [DOI] [PubMed] [Google Scholar]
- 9.Daniels R, Kurowski B, Johnson AE, Hebert DN. N-Linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 10.de Graaf M, Fouchier RAM. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vleeschauwer A, Atanasova K, Van Borm S, van den Berg T, Rasmussen TB, Uttenthal Å, Van Reeth K. Comparative pathogenesis of an avian H5N2 and a swine H1N1 influenza virus in pigs. PLoS ONE. 2009;4:e6662. doi: 10.1371/journal.pone.0006662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, Nolte DL, Ji G, He H. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS ONE. 2011 doi: 10.1371/journal.pone.0017212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, Zhang Y, Shinya K, Deng G, Jiang Y, Li Z, Guan Y, Tian G, Li Y, Shi J, Liu L, Zeng X, Bu Z, Xia X, Kawaoka Y, Chen H. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009;5:e1000709. doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs AJ, Armstrong JS, Downie JC. From where did the 2009 'swine-origin' influenza A virus (H1N1) emerge? Virol J. 2009;6:207. doi: 10.1186/1743-422X-6-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002;7:310–322. [PubMed] [Google Scholar]
- 16.Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4:9 pp
- 17.He L, Zhao G, Zhong L, Liu Q, Duan Z, Gu M, Wang X, Liu X, Liu X. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol. 2013;158:2531–2541. doi: 10.1007/s00705-013-1771-y. [DOI] [PubMed] [Google Scholar]
- 18.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Liu X, Li S, Guo X, Yang Y, Jin M. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J Virol. 2012;86:13879. doi: 10.1128/JVI.02726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang J, Bae SE. Comparative co-evolution analysis between the HA and NA genes of influenza A virus. Virology (Auckl) 2018;9:1178122x18788328. doi: 10.1177/1178122X18788328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi HJ, Gupta R. Eukaryotic glycosylation: online methods for site prediction on protein sequences. Methods Mol Biol. 2015;1273:127–137. doi: 10.1007/978-1-4939-2343-4_9. [DOI] [PubMed] [Google Scholar]
- 22.Karasin AI, Brown IH, Carman S, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasin AI, West K, Carman S, Olsen CW. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol. 2004;42:4349–4354. doi: 10.1128/JCM.42.9.4349-4354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim P, Jang YH, Kwon SB, Lee CM, Han G, Seong BL. Glycosylation of hemagglutinin and neuraminidase of influenza A virus as signature for ecological spillover and adaptation among influenza reservoirs. Viruses. 2018;10:183. doi: 10.3390/v10040183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klenk HD, Rott R. Cotranslational and posttranslational processing of viral glycoproteins. Curr Top Microbiol Immunol. 1980;90:19–48. doi: 10.1007/978-3-642-67717-5_2. [DOI] [PubMed] [Google Scholar]
- 26.Klenk HD, Wagner R, Heuer D, Wolff T. Importance of hemagglutinin glycosylation for the biological functions of influenza virus. Virus Res. 2002;82:73–75. doi: 10.1016/s0168-1702(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 27.Kosik I, Yewdell JW. Influenza hemagglutinin and neuraminidase: Yin(-)Yang proteins coevolving to thwart immunity. Viruses. 2019;11:346. doi: 10.3390/v11040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon TY, Lee SS, Kim CY, Shin JY, Sunwoo SY, Lyoo YS. Genetic characterization of H7N2 influenza virus isolated from pigs. Vet Microbiol. 2011;153:393–397. doi: 10.1016/j.vetmic.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Lee JH, Pascua PN, Song MS, Baek YH, Kim CJ, Choi HW, Sung MH, Webby RJ, Webster RG, Poo H, Choi YK. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J Virol. 2009;83:4205–4215. doi: 10.1128/JVI.02403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipatov AS, Kwon YK, Sarmento LV, Lager KM, Spackman E, Suarez DL, Swayne DE. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS Pathog. 2008;4:e1000102. doi: 10.1371/journal.ppat.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancera Gracia JC, Van den Hoecke S, Saelens X, Van Reeth K. Effect of serial pig passages on the adaptation of an avian H9N2 influenza virus to swine. PLoS ONE. 2017;12:e0175267. doi: 10.1371/journal.pone.0175267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCauley J, Bye J, Elder K, Gething MJ, Skehel JJ, Smith A, Waterfield MD. Influenza virus haemagglutinin signal sequence. FEBS Lett. 1979;108:422–426. doi: 10.1016/0014-5793(79)80578-5. [DOI] [PubMed] [Google Scholar]
- 34.Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernández R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovão NS, Lozano-Dubernard B, Rambaut A, van Bakel H, García-Sastre A (2016) Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5 [DOI] [PMC free article] [PubMed]
- 35.Mine J, Uchida Y, Sharshov K, Sobolev I, Shestopalov A, Saito T. Phylogeographic evidence for the inter- and intracontinental dissemination of avian influenza viruses via migration flyways. PLoS ONE. 2019;14:e0218506. doi: 10.1371/journal.pone.0218506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki-Horimoto K, Muramoto Y, Kawaoka Y. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16:1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 39.Parvin JD, Moscona A, Pan WT, Leider JM, Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986;59:377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci. 1999;96:1651. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell CJ, Webster RG. The genesis of a pandemic influenza virus. Cell. 2005;123:368–371. doi: 10.1016/j.cell.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Saavedra-Montanez M, Vaca L, Ramirez-Mendoza H, Gaitan-Peredo C, Bautista-Martinez R, Segura-Velazquez R, Cervantes-Torres J, Sanchez-Betancourt JI. Identification and genomic characterization of influenza viruses with different origin in Mexican pigs. Transbound Emerg Dis. 2019;66:186–194. doi: 10.1111/tbed.12998. [DOI] [PubMed] [Google Scholar]
- 43.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 45.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 46.Su S, Qi WB, Chen JD, Cao N, Zhu WJ, Yuan LG, Wang H, Zhang GH. Complete genome sequence of an avian-like H4N8 swine influenza virus discovered in southern China. J Virol. 2012;86:9542. doi: 10.1128/JVI.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun H, Kaplan BS, Guan M, Zhang G, Ye J, Long LP, Blackmon S, Yang CK, Chiang MJ, Xie H, Zhao N, Cooley J, Smith DF, Liao M, Cardona C, Li L, Wang GP, Webby R, Wan XF. Pathogenicity and transmission of a swine influenza A(H6N6) virus. Emerg Microbes Infect. 2017;6:e17. doi: 10.1038/emi.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 50.Tian H, Zhou S, Dong L, Van Boeckel TP, Cui Y, Newman SH, Takekawa JY, Prosser DJ, Xiao X, Wu Y, Cazelles B, Huang S, Yang R, Grenfell BT, Xu B. Avian influenza H5N1 viral and bird migration networks in Asia. Proc Natl Acad Sci. 2015;112:172. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venter M, Treurnicht FK, Buys A, Tempia S, Samudzi R, McAnerney J, Jacobs CA, Thomas J, Blumberg L. Risk of human infections with highly pathogenic H5N2 and low pathogenic H7N1 avian influenza strains during outbreaks in Ostriches in South Africa. J Infect Dis. 2017;216:S512–S519. doi: 10.1093/infdis/jix018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visher E, Whitefield SE, McCrone JT, Fitzsimmons W, Lauring AS. The mutational robustness of influenza A virus. Plos Pathog. 2016;12:e1005856–e1005856. doi: 10.1371/journal.ppat.1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vittecoq M, Grandhomme V, Simon G, Herve S, Blanchon T, Renaud F, Thomas F, Gauthier-Clerc M, van der Werf S. Study of influenza A virus in wild boars living in a major duck wintering site. Infect Genet Evol. 2012;12:483–486. doi: 10.1016/j.meegid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Wade A, Jumbo SD, Zecchin B, Fusaro A, Taiga T, Bianco A, Rodrigue PN, Salomoni A, Kameni JMF, Zamperin G, Nenkam R, Foupouapouognigni Y, Abdoulkadiri S, Aboubakar Y, Wiersma L, Cattoli G, Monne I. Highly pathogenic avian influenza A(H5N8) virus, Cameroon, 2017. Emerg Infect Dis. 2018;24:1367–1370. doi: 10.3201/eid2407.172120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang N, Zou W, Yang Y, Guo X, Hua Y, Zhang Q, Zhao Z, Jin M. Complete genome sequence of an H10N5 avian influenza virus isolated from pigs in central China. J Virol. 2012;86:13865–13866. doi: 10.1128/JVI.02687-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webster RG. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Arch Virol Suppl. 1997;13:105–113. doi: 10.1007/978-3-7091-6534-8_11. [DOI] [PubMed] [Google Scholar]
- 58.Weller CB, Cadmus KJ, Ehrhart EJ, Powers BE, Pabilonia KL. Detection and isolation of Influenza A virus subtype H1N1 from a small backyard swine herd in Colorado. J Vet Diagn Invest. 2013;25:782–784. doi: 10.1177/1040638713503655. [DOI] [PubMed] [Google Scholar]
- 59.Worobey M, Han GZ, Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci USA. 2014;111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Bao L, Deng W, Zhu H, Li F, Chen T, Lv Q, Yuan J, Xu Y, Li Y, Yao Y, Gu S, Yu P, Chen H, Qin C. Rapid adaptation of avian H7N9 virus in pigs. Virology. 2014;452–453:231–236. doi: 10.1016/j.virol.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Yang Y, Halloran ME, Sugimoto JD, Longini IM., Jr Detecting human-to-human transmission of avian influenza A (H5N1) Emerg Infect Dis. 2007;13:1348–1353. doi: 10.3201/eid1309.07-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zecchin B, Minoungou G, Fusaro A, Moctar S, Ouedraogo-Kabore A, Schivo A, Salviato A, Marciano S, Monne I. Influenza A(H9N2) virus, Burkina Faso. Emerg Infect Dis. 2017;23:2118–2119. doi: 10.3201/eid2312.171294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang G, Kong W, Qi W, Long L-P, Cao Z, Huang L, Qi H, Cao N, Wang W, Zhao F, Ning Z, Liao M, Wan X-F. Identification of an H6N6 swine influenza virus in southern China. Infect Genet Evol. 2011;11:1174–1177. doi: 10.1016/j.meegid.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017;45:D466–d474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]