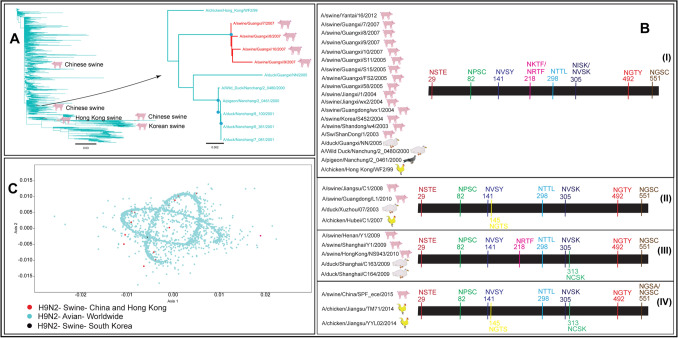
Fig. 3.
A Neighbour-joining tree of 6323 full-length HA nucleotide sequences of avian and swine H9N2 viruses determined multiple spillover events from avian reservoirs to swine. The red nodes represented avian to swine transmission of H9N2 viruses in China and Hong Kong, while black nodes represented avian to swine spillover in South Korea. The cyan nodes represented the circulation of H9N2 viruses among avian species globally. The blue dots at the nodes in the subtree represented > 90% bootstrap support. B Four patterns of glycosylation sites in swine and avian H9N2 viruses suggested multiple introductions and evolutionary trajectories of H9N2 viruses in swine in Southeast Asia. C The PCA plot represented multiple spillover events of H9N2 viruses in swine