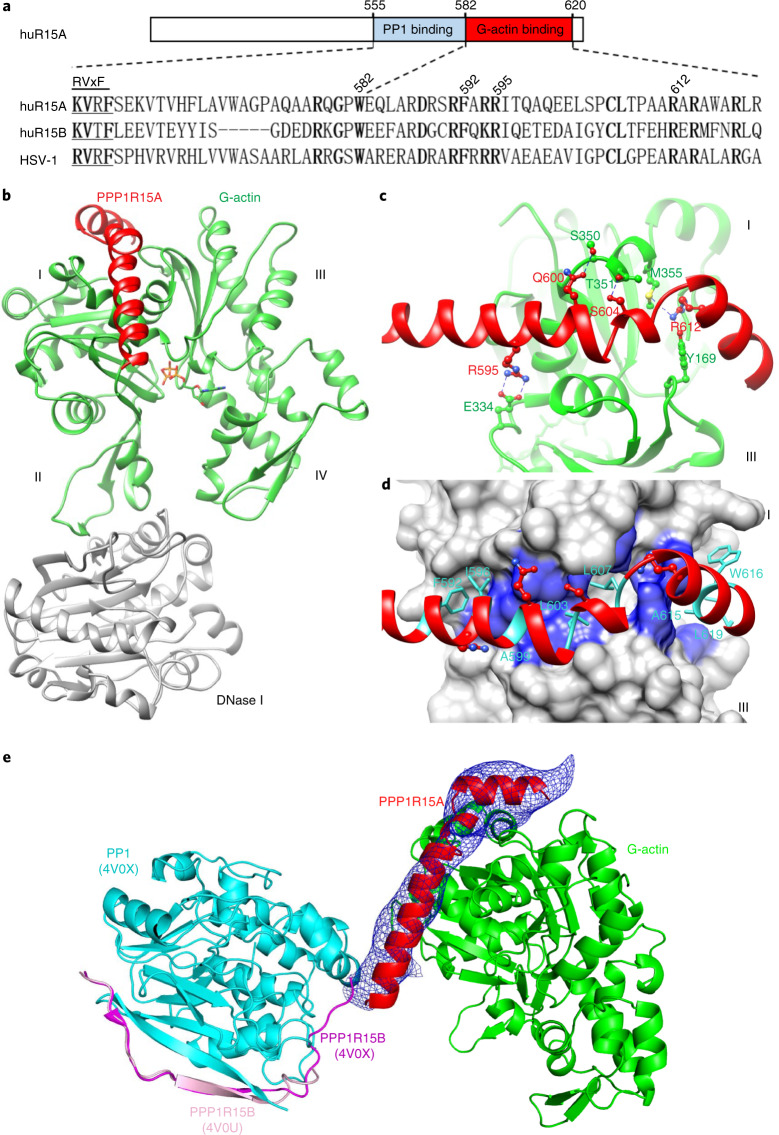
Fig. 1. PPP1R15A engages the barbed end of G-actin.
a, Schema of human PPP1R15A. The PP1- and G-actin-binding segments are marked. Beneath is an alignment of human PPP1R15A (huR15A), human PPP1R15B (huR15B) and the regulatory subunit of herpes simplex virus (HSV-1). Conserved residues are in bold. The ‘RVxF’ motif, common to many PPP1Rs, is noted. Numbering refers to human PPP1R15A. b, Ribbon diagram of the overall crystallographic structure of the PPP1R15A–G-actin–DNase I complex at a resolution of 2.55 Å. Actin domains are numbered (I–IV). c, Close-up view of hydrogen-bonding interactions between PPP1R15A (red) and G-actin (green). The density map for PPPR15A is shown in Extended Data Fig. 1. d, Hydrophobic interactions of G-actin (blue surface) with the indicated PPP1R15A residues (side chains as turquoise sticks). e, Model of a tripartite holophosphatase constructed by aligning the PPP1R15A–G-actin–DNase I complex (above) and the binary PP1G–PPP1R15B complex (PDB 4V0X) to the low-resolution PP1G–PPP1R15B–G-actin complex (PDB 4V0U); the former via G-actin and the latter via PP1. Shown is the PP1G–PPP1R15B from PDB 4V0X (with a root mean squared deviation (r.m.s.d.) of 0.318 Å between the 290 Cα pairs of PP1c) and the PPP1R15A–G-actin from the PPP1R15A–G-actin–DNase I complex (with r.m.s.d. of 0.626 Å between 343 Cα pairs of actin). DNase I is omitted for clarity. Note the proximity of the C terminus of PPP1R15B and the N terminus of PPP1R15A (consisting of the same residue of both orthologs: PPP1R15B Trp 662 and PPP1R15A Trp 582) in the two binary complexes. The previously unaccounted density in the barbed end of G-actin (displayed as an average difference electron density from PDB 4V0U and shown as a blue mesh) accommodates the actin-binding helices of PPP1R15A (red) from the aligned PPP1R15A–G-actin–DNase I complex.