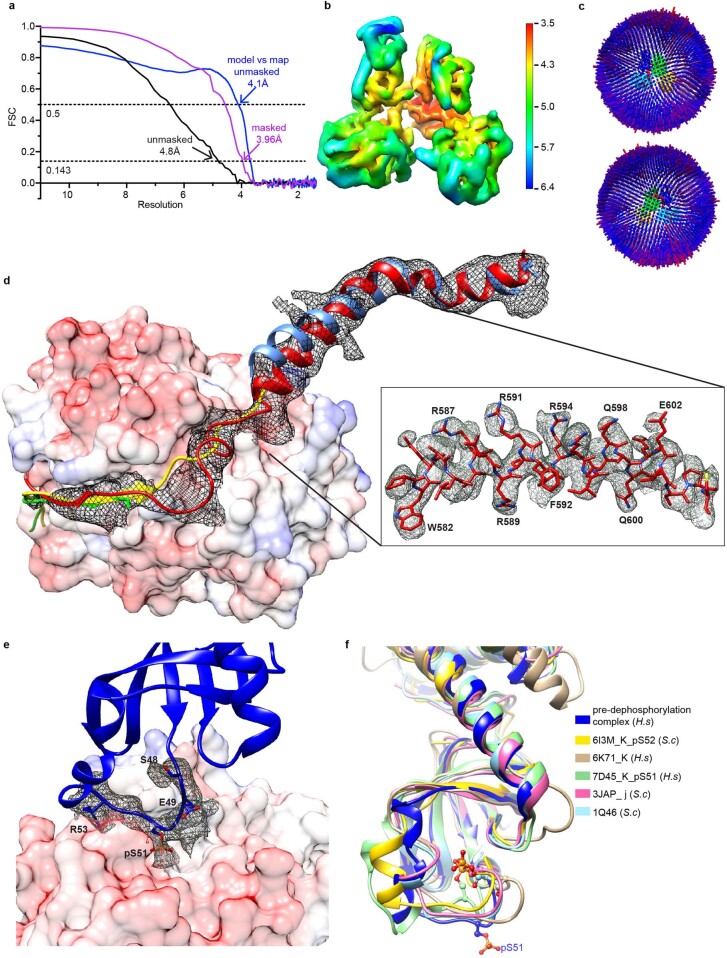
Extended Data Fig. 3. Further about the cryo-EM structure of the eIF2αP pre-dephosphorylation complex.
(a) The Fourier shell correlation (FSC) curves for masked (pink, produced using cryoSPARC), unmasked (black, produced using cryoSPARC) maps, and the curve for the unmasked model and map correlation (blue, produced in Phenix.real_space_refine) are shown. The resolutions at which FSC for masked map drops below 0.143 and model map correlation drops below 0.5 are indicated. (b) The 3D density map from non-uniform refinement in cryoSPARC is depicted by local resolution on a colour scale shown on the right. (c) Angular distribution plots of particles that contributed to the final map are viewed from the front and the back of the density map. The number of particles with respective orientations are represented by length and coloured cylinders (from blue to red). In the centre, the cryo-EM map of the pre-dephosphorylation complex is coloured and angled the same as in Fig. 4a. (d) Superposition of the GS-FSC density map for PPP1R15A in the cryo-EM pre-dephosphorylation complex (red), PPP1R15A from the crystal structure of its binary complex with PP1A (PDB 4XPN, green) and PPP1R15B from the crystal structure of its binary complex with PP1G (PDB 4V0X, yellow) (both aligned by PP1c) and PPP1R15A from the crystal structure of its binary complex with G-actin/DNase I (here, light blue, aligned by its G-actin). Inset is the density modified map of PPP1R15A581–606 from the Cryo-EM pre-dephosphorylation complex. (e) Density modified map for the eIF2αP substrate loop (residues 48-53) on the surface of PP1A, from the cryo-EM structure (eIF2α Arg52 sidechain was not modelled as there is no corresponding density and eIF2α Leu50 sidechain is omitted for clarity). (f) Alignment of human (H.s) eIF2α Ser51 loop from the pre-dephosphorylation complex with the structure of isolated yeast (S.c) eIF2α (PDB 1Q46), eIF2αP from human (H.s) (PDB 7D45 and 6K71) and yeast (PDB 6I3M) eIF2(αP)/eIF2B complexes and eIF2α from the yeast 48 S preinitiation complex (PDB 3JAP). PDB entry, chain and species are indicated. Note the unusual outward-facing orientation of phosphorylated Ser51 (pS51) from the pre-dephosphorylation complex.