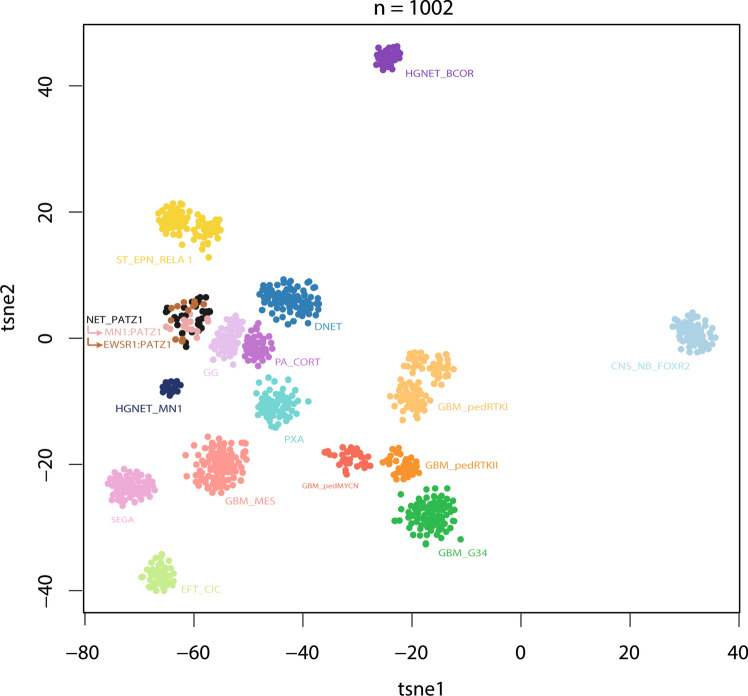
Fig. 1.
t-distributed stochastic neighbor embedding (tSNE) clustering of DNA methylation patterns of 60 NET-PATZ1 tumors alongside 942 in-house reference samples representing 15 other low- and high-grade glial and glioneuronal tumor types, using the 10,000 most variably methylated probes. NET-PATZ1 forms a distinct ‘island’. CNS_NB_FOXR2 CNS neuroblastoma with FOXR2 activation; DNET dysembryoplastic neuroepithelial tumor; EFT_CIC CNS ewing sarcoma family tumor with CIC alteration; GBM_G34 glioblastoma, H3.3 G34 mutant; GBM_MES glioblastoma, subclass mesenchymal; GBM_pedMYCN pediatric-type glioblastoma, subclass MYCN; GBM_pedRTKI pediatric-type glioblastoma, subclass RTKI; GBM_pedRTKII pediatric-type glioblastoma, subclass RTK II; GG ganglioglioma; HGNET_BCOR CNS high-grade neuroepithelial tumor with BCOR alteration; HGNET_MN1 CNS high-grade neuroepithelial tumor with MN1 alteration; NET_PATZ1 neuroepithelial tumor with PATZ1 fusion; PA_CORT hemispheric pilocytic astrocytoma; PXA pleomorphic xanthoastrocytoma; SEGA subependymal giant cell astrocytoma; ST_EPN_RELA supratentorial ependymoma, RELA fused