Figure 5. uORF Start Codon Context Sequence and Upstream Near Cognate Start Codons Contribute to Polyamine Control of HOL1 mRNA Translation.
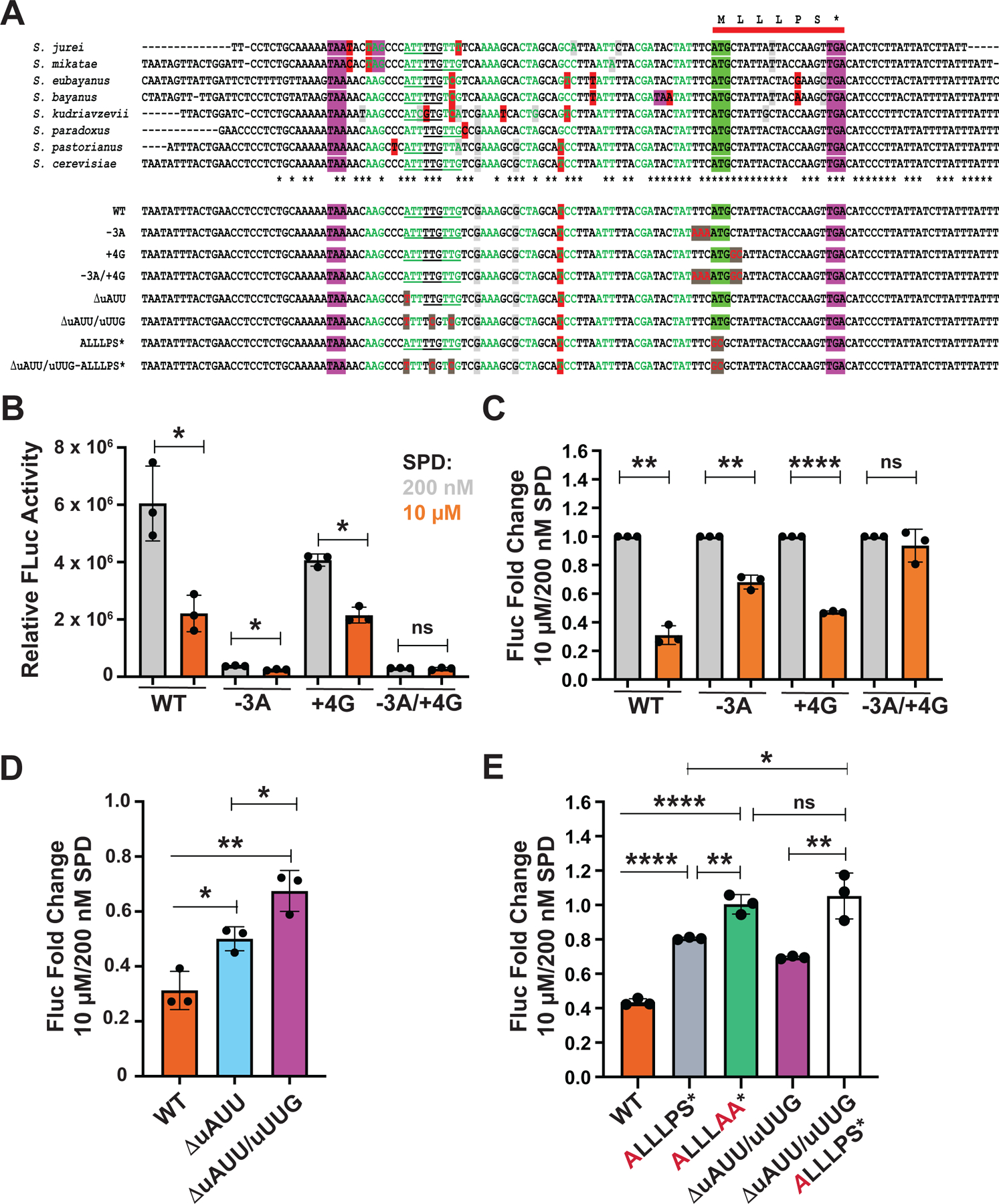
(A) (Upper panel) Multiple sequence alignment of a segment of HOL1 encoding the mRNA leader flanking the conserved uORF (red bar) from eight Saccharomyces species. The uORF start codon is highlighted in green and stop codons are highlighted in purple. The sequence upstream of the uORF is colored to highlight codons inframe with the uORF; nucleotide changes that alter the encoded amino acid are highlighted in red, while silent nucleotide changes that create synonymous codon substitutions are highlighted in light gray. Upstream inframe near cognate start codons are underlined; asterisks beneath alignment denote the positions that are conserved in all aligned sequences.
(Lower panel) Sequences of WT and mutant versions of S. cerevisiae HOL1 mRNA leader. Mutated residues are colored red and highlighted in dark gray.
(B) Relative HOL1-Fluc expression from the indicated WT or mutant reporters in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium supplemented with 200 nM (gray) or 10 μM (orange) SPD. Data were normalized to total protein. Error bars denote SD; *p < 0.05 (Student’s two-tailed t test; n = 3, assayed in duplicate).
(C) Data from (B) normalized to the 200 nM SPD controls. Error bars denote SD; **p < 0.01; ****p < 0.0001; ns, not significant (Student’s two-tailed t test; n = 3).
(D–E) Fold change in relative HOL1-Fluc expression from the indicated WT or mutant reporters in spe1 spe2 strain Y362 grown in polyamine-depleted SD medium supplemented with 200 nM or 10 μM SPD. Data were normalized to total protein and then to each 200 nM SPD control. Error bars denote SD; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (Student’s two-tailed t test; n = 3, assayed in duplicate).
