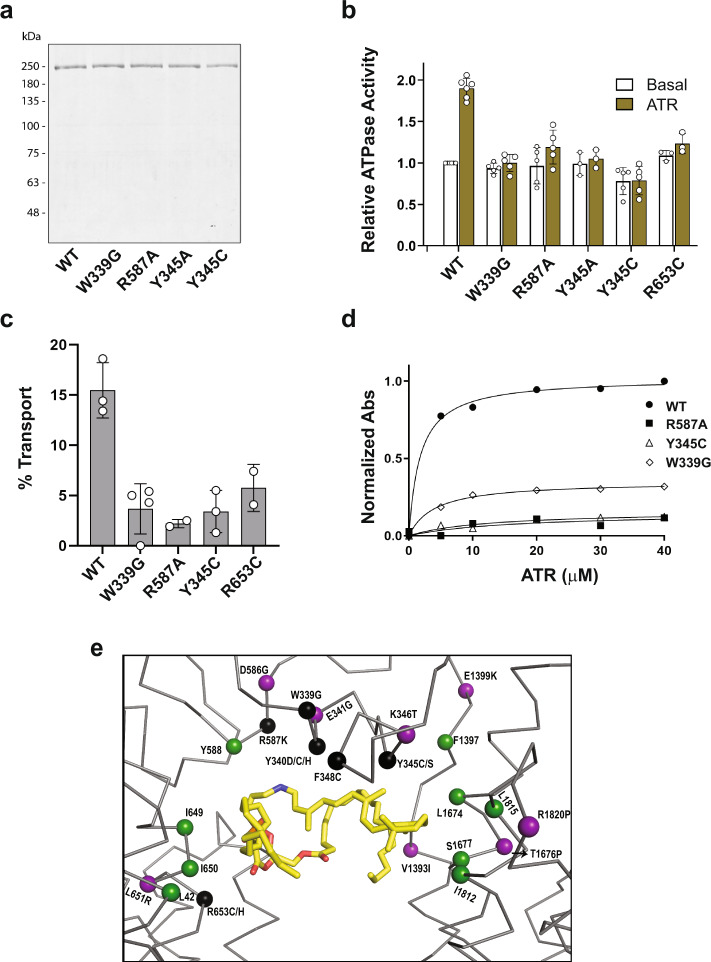
Fig. 8. Purification and functional characterization of ABCA4 variants with amino acid substitutions in residues involved in substrate binding.
a Representative coomassie blue stained gel of wild-type ABCA4 (WT) and variants expressed in HEK293T cells and purified by immunoaffinity chromatography. Data reproduced in three independent experiments. b ATPase activity of the ABCA4 variants in the absence (Basal) and presence of 40 µM all-trans retinal (ATR) to generate N-Ret-PE. Activity, normalized to WT basal activity, is expressed as the mean ± SD for n ≥ 3 independent experiments. P values between basal and ATR ATPase activities: WT = 0.001 (n = 5); W339G = 0.09 (n = 5); R587A = 0.009 (n = 5); Y345A = 0.07 (n = 3); Y345C = 0.82 (n = 5); R653C = 0.125 (n = 3) (two-tailed, paired Student T test). Data for R653C is from Garces et al.21 c % ATP-dependent transport of N-Ret-PE for samples treated with 1 mM ATP for 1 h. Data expressed as a mean ± SD for n = 3 (WT, Y345C) and n = 4 (W339G), and a range of values for n = 2 (R587A, R653C) independent experiments. d Binding of N-Ret-PE as a function of ATR concentration. WT ABCA4 had an apparent Kd of 1.7 ± 0.3 µM, R587A had an apparent Kd = 16.8 ± 24.6 µM, Y345C had a Kd of 12.6 ± 7.3 µM, and W339G had a Kd of 4.0 ± 0.5. Source data are provided as a Source Data file. e Location of key residues surrounding of the binding pocket. Protein is shown as ribbon and key residues as spheres. N-Ret-PE is shown as yellow sticks. Purple: Reported disease-associated variants in the vicinity of the binding pocket that do not directly interact with N-Ret-PE. Black: Disease-associated mutants that directly interact with N-Ret-PE. Green: Residues shown in our studies to interact with N-Ret-PE, but have not yet been reported to cause STGD1.