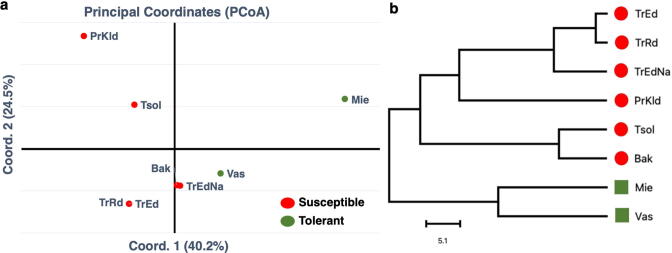
Fig. 6.
Identification and analysis of Single Nucleotide Variants (SNVs) located in the genomic sequence of pectin related hub-genes of eight sweet cherry cultivars. (a) Principal Coordinates Analysis (PCoA) and (b) Hierarchical clustering (Euclidean distance metric/spearman correlation method). Abbreviation of the cultivars: Bakirtzeika (Bak), Mieza (Mie), Proimo Kolindrou (PrKld), Tragana Edessis (TrEd), Tragana Edessis Naousis (TrEdNa), Tragana Rodochoriou (TrRd), Tsolakeika (Tsol), Vasileiadi (Vas). Red and green indicate cracking-susceptible and cracking-tolerant cultivars, respectively. For details, see the Supplementary Table 4. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)