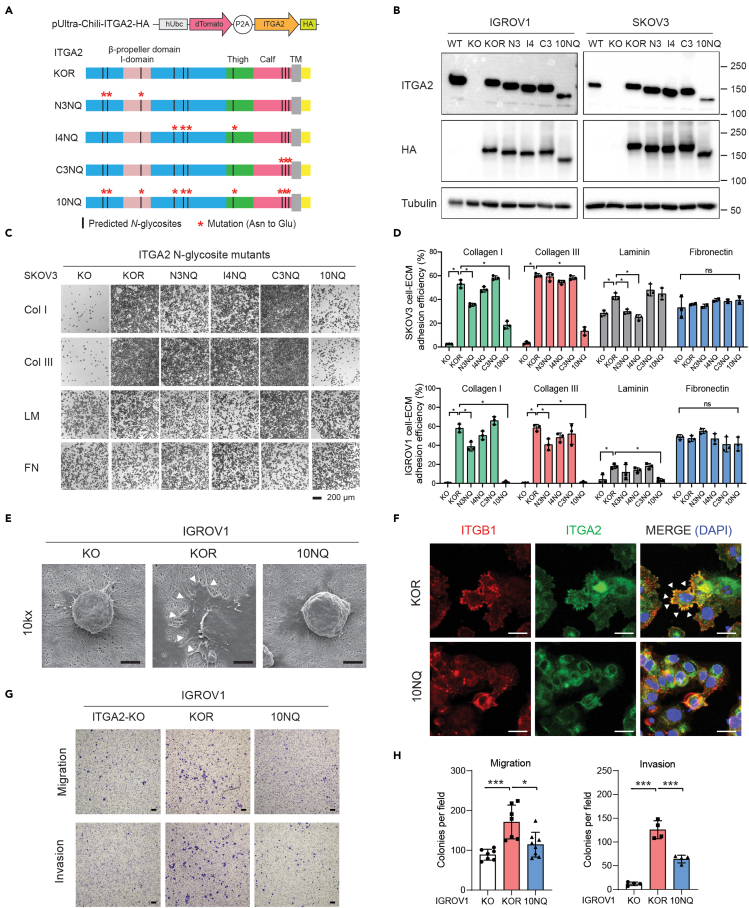
Figure 3.
N-glycosylation of ITGA2 affects cell-ECM adhesion
(A) Schematic representation of ITGA2 N-glycosites mutant constructs. Red asterisk indicates amino acid point mutation from Asn to Glu. The constructs are designed in KOR background (ΔITGA2 wildtype rescue), N3NQ (N105Q, N112Q, N343Q), I4NQ (N432Q, N460Q, N475Q, N699Q), C3NQ (N1057Q, N1074Q, N1081Q), and 10NQ (total N-glycosites mutation).
(B) Western blot with expression of mutant ITGA2 and C-terminal HA tag with corresponding mass shift.
(C) Static cancer cell-ECM adhesion assay. Representative images for ΔITGA2 (KO), KOR, and N-glycosite mutants (N3NQ, I4NQ, C3NQ, and 10NQ) cell adhesion to different ECM proteins.
(D) Bar charts show the quantification of the percentage of cell adhesion efficiency (mean ± SD) of SKOV3 and IGROV1 cells (ANOVA, ∗p < 0.05).
(E) Representative scanning electron microscope images with cellular adherence of IGROV1 KO, KOR, and 10NQ cells on 1.5% collagen-coated inserts. White arrows indicate membrane protrusions. Scale bar 5 μm.
(F) Immunofluorescence images of KOR and 10 NQ mutant cells co-stained with anti-HA (ITGA2-HA), Golgi marker (GM130) and nucleus (DAPI). Scale bar 20 μm.
(G) Representative 24h cell migration assay through 8 μm hanging inserts and 48h invasion assay with collagen-precoated inserts. Scale bar 100 μm.
(H) Bar charts show the mean ± SD of the migrated or invaded cells per field (mean ± SD) (ANOVA, ∗p < 0.05).