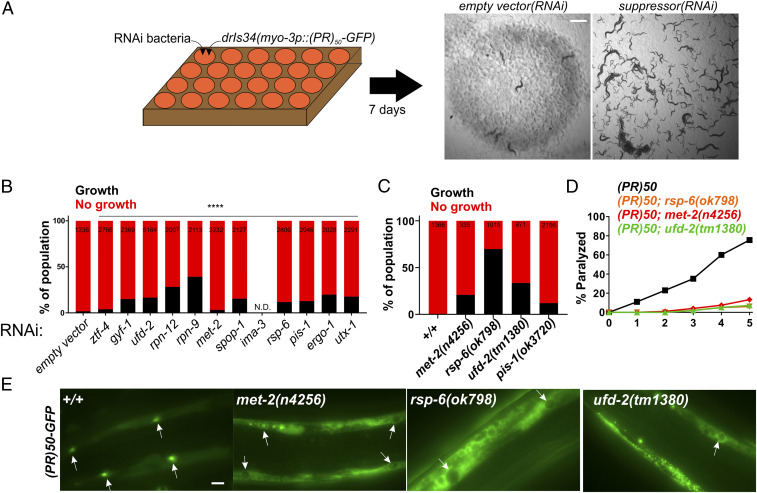
Fig. 1.
A genome-wide RNAi screen for suppressors of PR50 toxicity in C. elegans. (A) RNAi screening strategy. ∼30 hypochlorite isolated eggs from the drIs34 (myo-3p::PR50-GFP; myo-3p::mCherry) strain are seeded onto 24-well RNAi plates. After 7 d, each well was visually examined and scored relative to the empty vector(RNAi) control, which exhibits no growth and strong paralysis. Suppressors are identified as wells in which worms continue to grow and/or regain motility (gfp(RNAi) shown as an example). (B) COPAS quantification of worm length (time-of-flight [TOF]) in RNAi suppressors of PR50. Worms with a TOF >200 were placed in the “Growth” group while those with TOF <200 were placed in the “No growth” group. Data are presented as the percentage of the total population in either the “Growth” or “No growth” groups. The size of each population (N) is indicated at the top of each bar. ****P < 0.0001, Fisher’s exact test versus empty vector(RNAi). (C) Quantification of the percentage of animals expressing PR50 that reach the L4 stage (“Growth”) after 72 h at 25 °C. N is indicated at the top of each bar. (D) Age-dependent paralysis assay was performed with homozygous viable mutations in genes identified in the RNAi screen. n = 100 for each genotype. For each genotype versus WT, P < 0.0001, log-rank test with Bonferroni correction. (E) Localization of PR50-GFP in either the WT or indicated homozygous mutant background. Arrows point to muscle nucleus. In WT, PR50-GFP is primarily localized to nuclei but in n4256, ok798, and tm1380, PR50-GFP is largely excluded from the nucleus and accumulates in nonnuclear compartments. (Scale bar, 10 microns.)