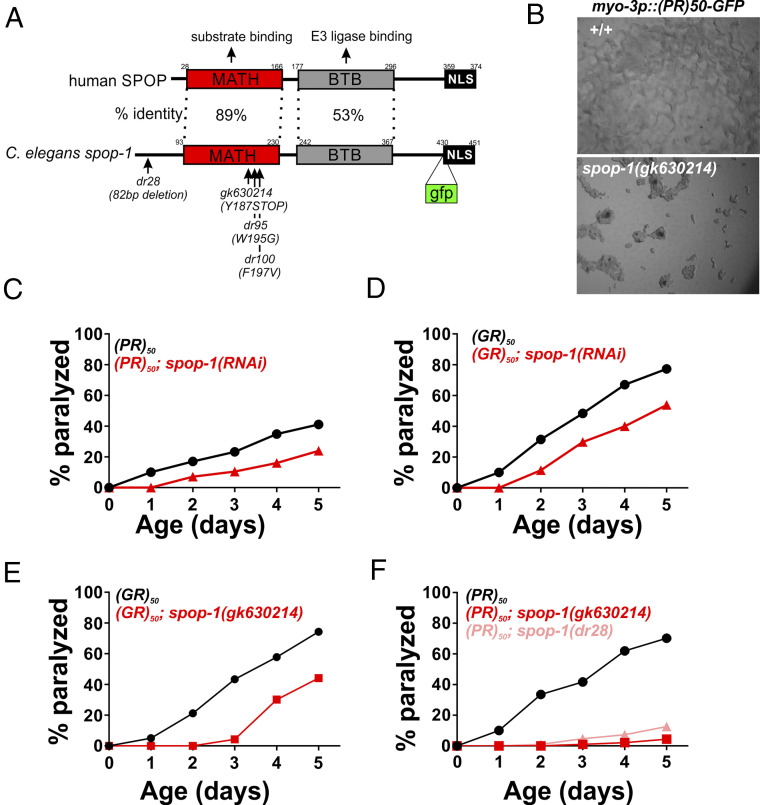
Fig. 2.
Loss-of-function mutations in the E3 ubiquitin ligase adaptor spop-1 suppress PR50 and GR50 toxicity in C. elegans. (A) Domain structure of the SPOP protein. The percent identity between the human and C. elegans domains are shown. The dr28 allele is a CRISPR-induced allele that deletes 82 bp within the second exon of spop-1. This shifts the reading frame of isoform 1 to encounter a premature stop codon and deletes that start ATG for isoform 2. Therefore, dr28 is a likely spop-1 null allele. The gk630214 allele is a spop-1 nonsense mutation isolated from the Million Mutation project. The gfp insertion site used to make the SPOP-1-GFP CRISPR allele is shown. (B) spop-1(gk630214) suppresses the developmental arrest phenotype of PR50-GFP. (C and D) spop-1(RNAi) suppresses the age-dependent paralysis phenotype of PR50-GFP (P = 0.000018) (C) and GR50-GFP (P = 0.0001) (D). (E and F) spop-1 alleles gk630214 and dr28 suppress PR50-GFP (P = 0.00000011) (E) and GR50-GFP (P < 0.0001) (F). For all paralysis assays, n = 100 animals for each genotype.