Figure 1. Senescent cells synthesize oxylipins.
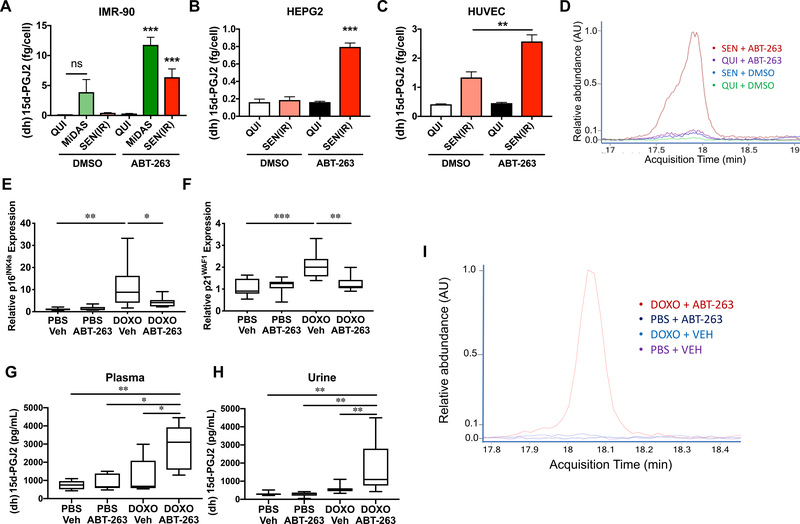
A-B. IMR-90 cells were proliferative in 10% serum, made quiescent by incubation for 3 days in 0.2% serum, or made senescent by treatment with 10 Gy X-rays (IR) or ethidium bromide to induce mitochondrial-dysfunction induced senescence (MiDAS). Lipids were extracted from proliferating (PRO – 10%), quiescent (QUI – 0.2%), IR-induced senescent (SEN(IR) – 10% or 0.2% serum), or mitochondrial dysfunction-associated senescent (MiDAS – 0.2%) cells and analyzed by liquid chromatography combined with mass spectrometry (LC-MS). Putative oxylipins (A) and lipid precursors (B) were detected in control and senescent cells. Heat maps indicate the averages of at least 3 experiments (* = p<0.05, 1-way ANOVA). C. RNA was isolated from QUI, SEN(IR), MiDAS and vector or RAS-V12 expressing [SEN(RAS)] cells, reverse transcribed, and mRNAs encoding oxylipin synthesis genes were measured by quantitative (q) PCR. Heat maps indicate the averages of 3 experiments (* = p<0.05, 1-way ANOVA). D. p16–3MR mice were given a single dose (10 mg/kg) of doxorubicin (DOXO) or phosphate-buffered saline (PBS) by intraperitoneal (i.p.) injection. After 5 days, mice were given GCV (25 mg/kg) or vehicle by i.p. injection for 5 consecutive days. Livers were harvested on day 10 and analyzed for oxylipin synthase mRNAs by qPCR. Heat maps indicate averages of at least 5 mice (* = p<0.05, 1-way ANOVA). E. IMR-90 fibroblasts were induced to senesce by MiDAS, IR, or RAS-V12 overexpression. 10 days after induction of senescence, conditioned media were collected and analyzed for PGD2 by ELISA. F-G. HEPG2 (F) and HUVEC (G) cells were induced to senesce by IR. Conditioned media were collected 10 days after IR and analyzed for PGD2 by ELISA. Data show means of at least 3 experiments (* = p<0.05, ** = p<0.01, *** = p<0.001, unpaired, 2-tailed t test). H-I. RNA from (C) was analyzed for expression of SLCO2A1 or PGDH by qPCR. J. HUVECs were induced to senesce by IR, and analyzed at Day 10 for SLCO2A1 and PGDH as in H and I. See also Figure S1 and Table S1.