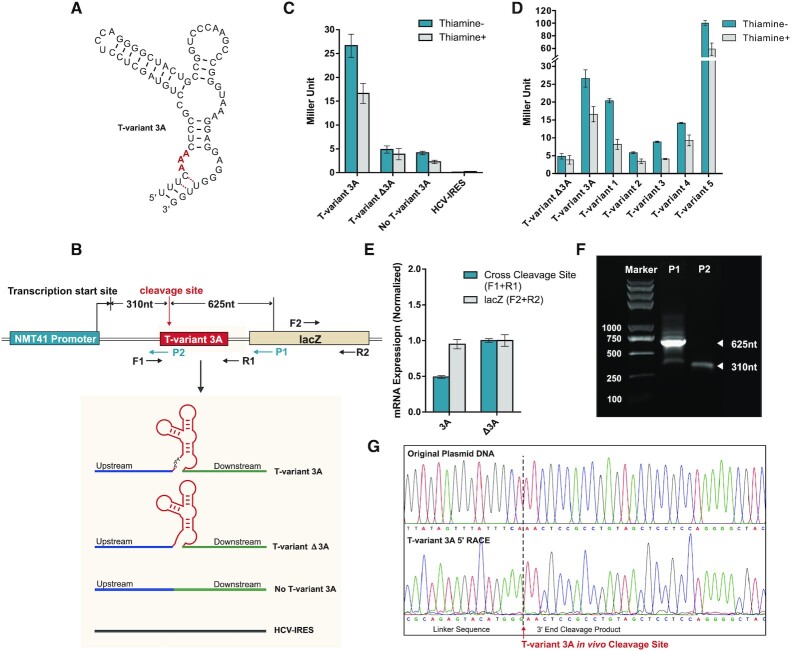
Figure 4.
Reporter constructs and T-variant in vivo catalytic activity. (A) Sequence and secondary structure of T-variant 3A. (B) The reporter plasmid constructs. The wild type T-variant 3A for 5′ RACE is located behind an NMT41 thiamine repressible promoter and in front of a lacZ reporter gene. The gene specific primers P1 and P2 were used to map the T-variant 3A in vivo cleavage site and transcription start sites respectively. The T-variant 3A cleavage site as determined in vitro is marked by the red arrow and the predicted P1 and P2 primer fragment sizes of cleaved and whole transcript shown. The control plasmid constructs for real-time PCR and Miller assay are shown in the grey box: In parallel with T-variant 3A (red box), a sequence containing only the peripheral sequence of the Perere-3 5′-UTR without the ribozyme (No T-variant 3A) or with an HCV-IRES were substituted into the position of the red box in the lacZ reporter. (C) Miller assay of lacZ reporter activity ± thiamine for T-variant 3A, no T-variant and HCV IRES plasmid constructs. Note the high levels of reporter gene expression for T-variant 3A relative to the controls when the ribozyme is active. (D) Miller assay of lacZ reporter activity ± thiamine for T-variant sequences 1 to 5, relative to the control constructs T-variants Δ3A and 3A. (E) Real-time PCR analysis of mRNA abundance of the lacZ RNA (F2 and R2 primers) relative to the Ampicillin internal reference, showing the level of mRNA abundance of lacZ in the wild type T-variant 3A remains stable after T-variant cleavage, compared to the controls. Error bars represent the standard deviation of three independent experiments. (F) Agarose gel of PCR product of 5′RACE; the P1 primer generates a 625 nt T-variant cleavage product. (G) Capillary electrophoresis sequencing of original plasmid DNA compared to the 5′ RACE of cleaved T-variant 3A in vivo, the T-variant 3A cleavage site is marked by the red arrow.