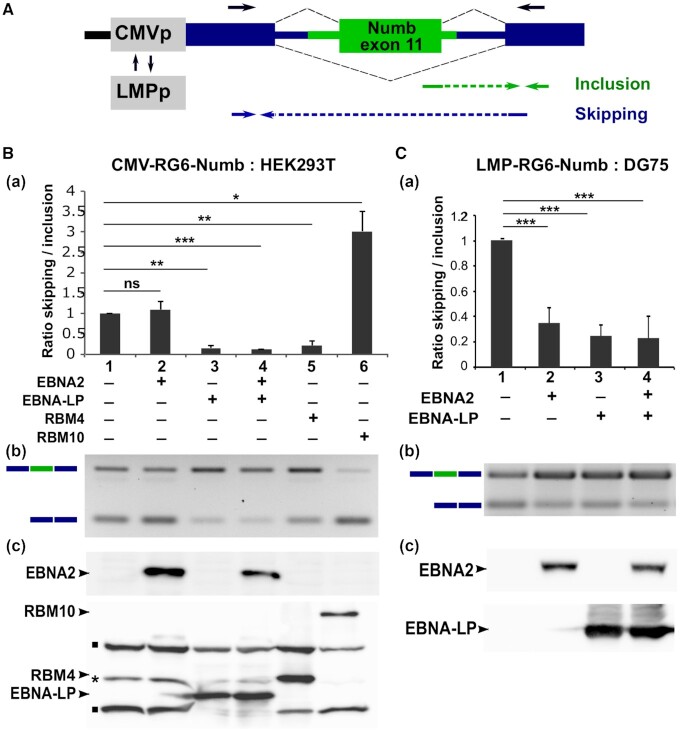
Figure 5.
modulation of Numb exon 11 alternative splicing by EBNA2 and EBNA-LP. (A) Schematic representation of the RG6-Numb reporter construct (32). The green box and lines represent the human NUMB exon 11 and 100/50 nucleotides of the flanking 5′/3′ intronic regions, respectively. Dark blue boxes and lines indicate constitutive exons and intronic sequences specific for the RG6 minigene (34). The reporter gene is under the transcriptional control of either the cytomegalovirus immediate early promoter (CMVp) or the EBV LMP viral promoter (LMPp). Arrows placed above the RG6-Numb schematic sequence indicate the location of primers used for semi-quantitative RT-PCR. Arrows placed below indicate the location of primers used for quantitative RT-PCR (RTqPCR) to amplify specifically the inclusive or skipping isoforms. HEK293T (B) or DG75 B cells (C) were transfected with the pCMV-RG6-Numb or pLMP-RG6-Numb reporter constructs respectively, together with expression plasmids coding for EBNA2, EBNA-LP, RBM4 or RBM10 as indicated in the Figures. Panels a: relative ratio between exon inclusion and skipping are estimated using quantitative RT-PCR. Error bars represent the standard deviation from three independent experiments. The P values of paired Student's t-test are reported as *P < 0.1; **P< 0.05; ***P< 0.001; ns, not significant. Panels b: agarose gel analysis of a representative semi-quantitative RT-PCR reaction using primers depicted in A (black arrows) from one of the three experiments used for the quantitative RT-PCRs. Panels c: representative western blot analysis of one of the three experiments. Upper panels: EBNA2 is revealed using an anti-EBNA2 mAb (PE2, abcam). Lower panels: RBM4, RBM10 and EBNA-LP being all tagged with the Flag epitope were revealed using anti-Flag M2 mAb (Sigma-Aldrich). N.B.: the RG6-Numb construct carries both dsRED and EGFP coding sequences that are alternatively put in frame depending on the inclusion or skipping of Numb exon 11 (34). A Flag epitope sequence placed immediately upstream of the first exon of RG6-Numb allows the detection of these two fusion proteins by anti-Flag M2 mAb as seen on the gel (bands indicated by a black square). The higher molecular weight band corresponds to the GFP in-frame fusion generated when Numb exon 11 is included, whereas the lower molecular weight band corresponds to the dsRED in-frame fusion generated when Numb exon 11 is skipped. * Unspecific band.